Figure 3. Certain properties of the smaller ligand’s complex correlate with increased likelihood of changing binding mode when elaborated.
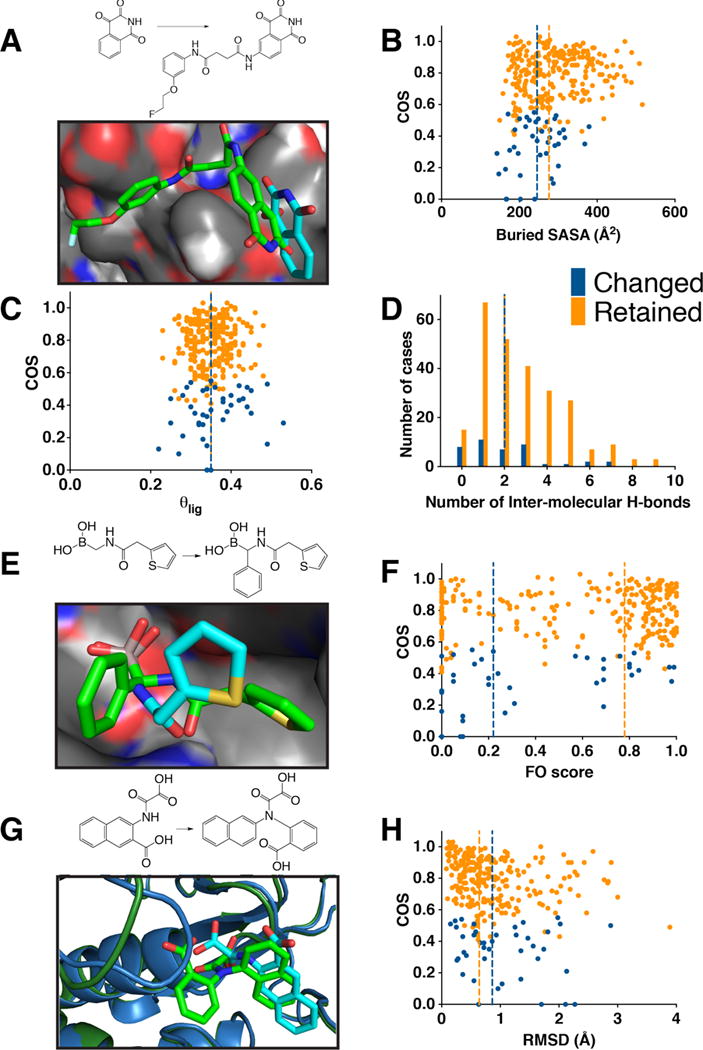
In each case, blue indicates cases in which the smaller ligand changes binding mode upon elaboration, and orange is used for cases in which the binding mode is preserved. Vertical lines indicate the medians of each distribution. (A) In this example of alternate binding modes, the smaller ligand (cyan, PDB ID 3deh) uses a very shallow binding mode on the surface of caspase-3; upon elaboration, the binding mode of the larger ligand retains the position of this structural element, but has reversed the relative orientation of the arrangement of the polar and non-polar sides of this fragment (green, PDB ID 3dek). (B) Compounds that change binding mode upon elaboration typically bury less solvent accessible surface area than compounds that retain their binding mode (p < 9×10−4). (C) Compounds that change binding mode upon elaboration do not bind with more shallow binding modes, which would correspond to higher θlig values (θlig is the fraction of the ligand’s SASA that remains exposed upon complexation). (D) Compounds that change binding mode upon elaboration typically have fewer intermolecular hydrogen bonds (p < 0.02), even though the median value is the same for this discrete variable. (E) In this example of alternate binding modes (cyan, PDB ID 1fsw; green, PDB ID 1my8), both β-lactamase inhibitors make identical hydrogen bonds using their boronic acid groups; upon addition of an extra phenyl ring, however, the amide linker flips over to position the thiophene in a very different location. (F) Compounds that change binding mode upon elaboration typically have lower FO scores (a measure of the extent to which the smaller ligand fills the larger ligand’s “binding energy hot spot”) than compounds that retain their binding mode (p < 5×10−4). (G) In this example of alternate binding modes, the smaller ligand forms stacking interactions with a phenylalanine sidechain in the binding site (cyan, PDB ID 1c84). Elaboration with a benzoic acid group pushes away this phenylalanine sidechain, and instead forms new hydrogen bonds that require the ligand to move within the binding site (green, PDB ID 1no6). Meanwhile, this larger ligand pushes away a loop that previously covered the binding site (left), which is primarily responsible for the RMSD difference between the two protein structures. (H) Compounds that change binding mode upon elaboration are more often accompanied by conformational rearrangement of the protein’s binding site, relative to compounds that retain their binding mode (p < 0.03).
