Figure 9. Fragments are particularly prone to alternate binding modes.
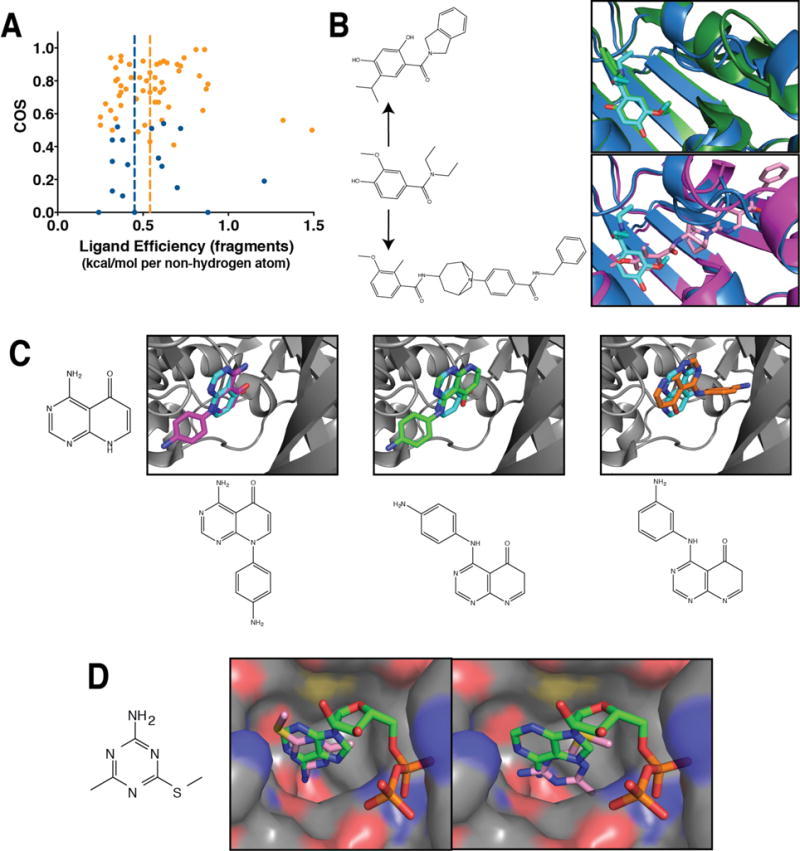
(A) Among fragment starting points, there is no statistically significant difference in ligand efficiency between those that change binding mode upon elaboration versus those that retain their binding mode (p < 0.3). (B) Fragment screening for ATP-competitive Hsp90 inhibitors yielded an initial hit (cyan, PDB ID 2×dl) that was elaborated into a more potent lead while perfectly preserving the binding mode (top, green, PDB ID 2xab). Separately, a high-throughput screen yielded a compound with related chemical structure that positions the corresponding ring in a completely different orientation from that of the fragment (bottom, pink, PDB ID 4awq). (C) The structure of the 4-amino-8H-pyrido[2,3-d]pyrimidin-5-one core compound was solved in complex with TGFBR1 (cyan, PDB ID 4×0m), and found to engage with the kinase hinge region through a specific set of hydrogen bonds. Elaborating with an anilino group at one position preserved the binding mode (left, magenta, PDB ID 4×2f), whereas substituting this anilino group at two other positions yielded two more distinct binding modes (middle, green, PDB ID 4×2g; right, orange, PDB ID 4×2j). (D) Fragment screening for ATP-competitive Hsp90 inhibitors led to 4-methyl-6-(methylsulfanyl)-1,3,5-triazin-2-amine. When this compound is co-crystallized with the protein (left, PDB ID 2wi2), it closely mimics the interactions of ADP. However, soaking the same compound into protein crystal yields a different binding mode (right, PDB ID 2wi3), which makes different interactions and offers distinct opportunities for optimization.
