Abstract
Sustainable and personally tailored materials production is an emerging challenge to society. Living organisms can produce and pattern an extraordinarily wide range of different molecules in a sustainable way. These natural systems offer an abundant source of inspiration for the development of new environmentally friendly materials production techniques. In this paper, we describe the first steps toward the 3-dimensional printing of bacterial cultures for materials production and patterning. This methodology combines the capability of bacteria to form new materials with the reproducibility and tailored approach of 3D printing systems. For this purpose, a commercial 3D printer was modified for bacterial systems, and new alginate-based bioink chemistry was developed. Printing temperature, printhead speed, and bioink extrusion rate were all adapted and customized to maximize bacterial health and spatial resolution of printed structures. Our combination of 3D printing technology with biological systems enables a sustainable approach for the production of numerous new materials.
Keywords: 3D printing, synthetic biology, Escherichia coli, alginate
The development of more sustainable materials production is an urgent need for our current society. Traditional materials production processes can utilize massive quantities of polluting chemicals, and too often the products are not naturally degradable.1 One newly emerging approach to solve this problem is the production of materials by bacteria, either synthetically modified2−4 or unmodified.5,6 Many bacteria can carry out advanced chemical reactions to produce materials, including amyloid-based adhesives,7 biobased electrical switches,8 microbially produced nacre,9 and a variety of bioplastics,10 under ambient conditions without using or producing toxic compounds. However, current microbial materials production techniques do not allow the generation of bespoke material structures in a reliable or reproducible way. To allow for the production of complex patterned biomaterials, we have coupled bacterial materials production with 3D printing technology.11
Additive manufacturing allows for the production of tailored products fulfilling individual needs and enables entrepreneurs and companies to produce small batches or only on-demand.12 A wide variety of 3D printing approaches have been developed for additive manufacturing of nonbiological materials, including stereolithography, selective laser sintering, electron beam melting (EBM), Laser Engineered Net Shaping, and PolyJet.13,14 The vast majority of current 3D printing techniques involve one or more steps that are deadly for cells. Therefore, new technologies have been developed for cellular printing with medical and biological applications including visualization, education, and transplantation.15−17 To date, these techniques are quite expensive (between 5000 and 200 000 USD18) and not yet well adapted for bacteria.
The area of biological printing is primarily dominated by three different technologies. Thermal inkjet bioprinting uses CT or MRI images as guides to deploy fluid dots layer by layer, which solidify to a gel after a short heating phase in the extruder.19 Direct write bioprinting utilizes print-heads made of syringes or needles that transport liquid bioink via mechanical or pneumatic systems and uses a low-melting-point scaffold material.20,21 Spheroid organ printing employs tiny tissue spheroids to assist the self-organizational and self-assembling character of real tissues.22 A few efforts have been made to apply these technologies to bacterial printing, but all current approaches suffer limitations of poor spatial resolution23 or require laborious clean-room fabrication of microstructures that shape the printed bacteria.24 To achieve a cost-efficient technology for bacterial printing that is compatible with incubation of printed bacteria at elevated temperatures, a new technology must be developed that allows high printing resolution without the requirement for cleanroom facilities.
Our newly developed microbial 3D printer can deposit bacteria cells in specific three-dimensional patterns for the ultimate goal of materials production. Our printing platform uses a modified commercially available 3D printer to extrude a mixture of bacteria and alginate that solidifies into a gel upon contact with a calcium chloride-treated printing surface. This combination of straightforward chemistry and easy, readily available technology enabled us to print reproducible 3-dimensional samples with high spatial resolution. The combination of our straightforward technique to print 3D microbial structures with the material-modifying properties of bacteria will result in high-resolution deposition of bacteria and the fabrication of spatially patterned materials.
Results and Discussion
3D Printer and Bioink
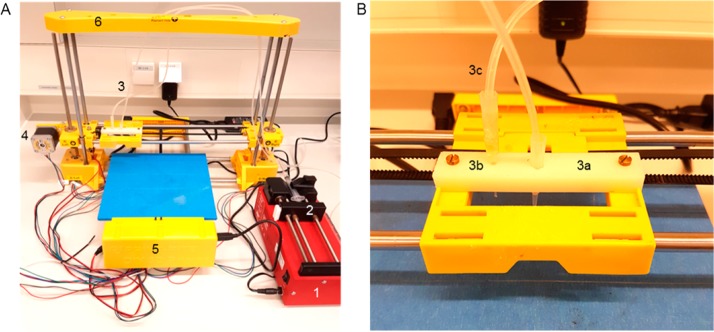
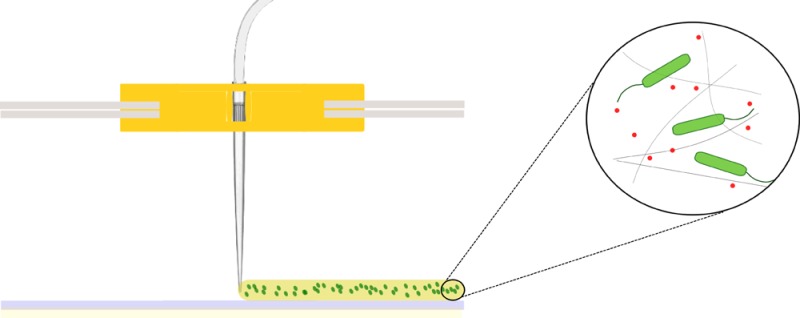
In order to create a straightforward bacterial 3D printer, multiple modifications were made to an inexpensive (300 USD) commercial 3D printer (Figure 1A). The extruder of the printer was removed and replaced with a pipet tip (Figure 1B, 3a) and system of tubing (Figure 1B, 3c). This alteration allows the liquid biological ink (“bioink”) to be transported under ambient temperatures that are amenable to microbes, rather than the elevated temperatures that are applied to melt plastic filament. A secondary pipet tip was affixed to the printhead (Figure 1B, 3b) to allow for rapid alternation between the deposition of different types of bioink. A syringe pump (Figure 1A, 1) was added to the system to generate continuous but adjustable flows of bioink through the sets of tubing into the pipet tips. Printed shapes are created through the flow of bioink through the movable printhead while it is in motion, the trajectory of which is programmably controlled by an external computer. The shape of printed objects can be created in silico through computer-aided design (CAD) software programs, then converted into printing instructions for the 3D printer using slicing and printer-specific software programs. These adaptations can be performed on all 3D printing systems that employ an accessible and removable extruder.
Figure 1.
Bacterial 3D printing system. (A) Overview of all bioprinter components. 1: syringe pump, 2: syringe filled with bioink, 3: extruder holder, 4: one of three step-motors for positioning, 5: breadboard and hardware of the printer, 6: frame of the printer. (B) Detailed view of the modified extruder. 3a: active pipet tip, 3b: secondary pipet tip for layering materials, 3c: tubing system.
A custom bioink was developed that would allow bacteria and chemical substrates for materials production to flow through the printhead in liquid state, then rapidly solidify upon contact with the printing surface to form a stably patterned shape. The bioink consists of live bacteria mixed with dissolved alginate. When the bioink is extruded onto a surface containing calcium ions during the printing process, cross-linking of the alginate molecules is triggered, forming a stable, biocompatible aerogel scaffold within seconds.25,26 In order to optimize the bioink composition, the alginate and calcium ion concentration were systematically varied (from 0.5% w/v to 6% w/v alginate; and 0.0087 mol/cm2 to 0.44 mol/cm2 CaCl2) and tested in the printing system. Insufficient concentrations of alginate and calcium ions resulted in poor gelation and low printing resolution, while excessive concentrations led to premature gelation of the bioink, blocking the pipet tip and preventing further printing. The optimal conditions were found to be 1 M CaCl2 and 2.5% w/v alginate. With our printer system, each printed layer can only contain materials that are present in the active syringe; the mixture of materials from two different syringes within one layer is not possible due to the rate of scaffold formation. However, a wide range of different types of materials is compatible with alginate polymerization and may be admixed in the active syringe for inclusion in individual printed layers.21,27
Printing Reproducibility and Resolution
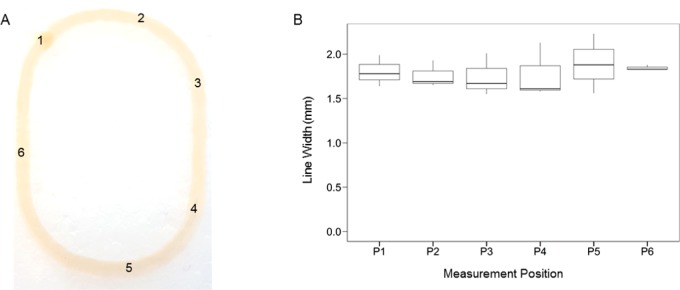
The reproducibility and consistency of the printing process was assessed by analyzing patterned monolayers that were printed using the optimized bioink. An elliptical form with two long straight lines on each side was chosen as the printing object in order to test the performance of the printer in fabricating both curved and straight-edged structures (Figure 2A, Supporting Information). Following printing, the width of each printed structure was measured at six different positions, sampling a variety of straight and curved portions. No statistically significant differences were seen among any of the widths measured at the same position in different prints (ANOVA, p-value: 0.964, CV%: 11.16) (Figure 2B). These data indicate that our printer and bioink can fabricate printed structures of varying shapes in a consistently uniform manner.
Figure 2.
Reproducible printing of alginate structures. (A) A representative printed elliptical structure. The numbers indicate the locations of the 6 measurement positions. (B) The distribution of measured line widths at 6 different positions within elliptical printed structures (n = 3). The tops and bottoms of the boxes represent the 75th and the 25th percentiles, respectively; the lines within the boxes are the median values; and the tops and bottoms of the vertical lines are the maximum and minimum values. No significant differences exist between the width distributions of any of the different positions (ANOVA, p-value: 0.964).
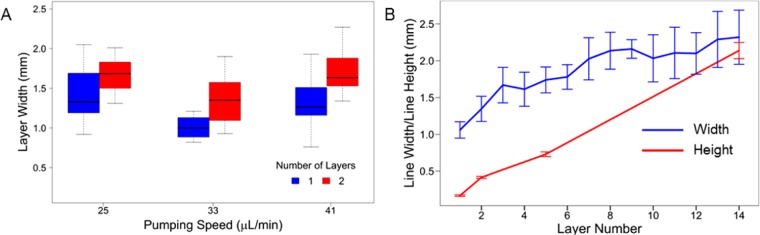
In order to maximize the printer resolution, a range of printing parameters was tested. The two most critical factors affecting the printed line width were found to be the extrusion rate of the syringe pump and the movement speed of the printhead, in agreement with previous work.28 A range of printhead movement speeds (100–500 mm/min) and syringe pump extrusion rates (17–50 μL/min) were applied to print straight lines of bioink. In general, increased printing resolution could only be achieved by adjusting both parameters in parallel: e.g., slower printhead movement speeds required slower syringe pump rates. Other less-critical factors contributing to printer resolution were the distance between the printhead and the printing surface, as well as the uniformity of the printing surface. The narrowest line width obtainable was 1.00 ± 0.15 mm, achieved with a printhead movement speed of 200 mm/min and a syringe pump extrusion rate of 33 μL/min (Figure 3A).
Figure 3.
Printing at millimeter-scale resolution in three dimensions. (A) The distribution of widths of single- and double-layered structures printed at different syringe pump speeds (n = 4). The printhead movement speed was 200 mm/min in all cases. The tops and bottoms of the boxes represent the 75th and the 25th percentiles, respectively; the lines within the boxes are the median values; and the tops and bottoms of the vertical lines are the maximum and minimum values. (B) The line width (blue) and line height (red) of printed structures containing up to 14 layers (n = 3). Error bars indicate the standard error.
The printer can be directed to deposit bioink directly on top of previously printed material to create multilayered structures. A second aerogel layer can be printed on top of a base layer at a range of different syringe pump extrusion rates, with no modification of the printing commands (Figure 3A). Fabrication of structures taller than two layers in height requires an increase of the z-position of the printhead by 0.15 mm/layer. Stacked layers of bioink are able to solidify due to interaction with calcium ions that have diffused from the printing surface up through the first printed layer(s). Each additional printed layer resulted in a fractional increase in the width of the final structure, due to the time required for the new layer to gelate (Figure 3A, B). No significant change in the final width was observed when the time between printing of successive layers was varied between 40 and 240 s (Student’s t test, p-value: 0.037, CV40s: 18.36, CV240s: 20.82), indicating that multilayered structures can be printed at different paces with no loss of resolution. The total time required to print a 14-layered ellipse with a pause of 40 s between printing successive layers was 15 min.
To characterize the spatial resolution of 3-dimensional printed structures, 14-layered elliptical structures were printed. The structures’ widths were measured following deposition of each layer. The heights of the structures were measured for only a subset of layers, since each height measurement required removal of the gel from the printing surface, halting the printing process. The average line width increased significantly but incrementally for the first six layers, with an average increase of 0.14 ± 0.01 mm per layer between layers 1 and 6 (Figure 3B). Following the sixth layer, the line widths approached a plateau; no significant differences were observed between the line widths of any of the layers between layers 6 and 14 (ANOVA + Tukey post hoc test, p-value: 0.995). The height of the printed material was observed to increase continually, by an average of 0.16 ± 0.02 mm per layer. The final 14-layered structures were 2.14 ± 0.11 mm in height, with a width of 2.32 ± 0.37 mm. These measurements indicate that our printing system is capable of fabricating 3-dimensional structures at submillimeter-scale precision in all dimensions. Further improvements in resolution may be possible by rebuilding our system using a commercial 3D printer employing more accurate printhead positioning.15
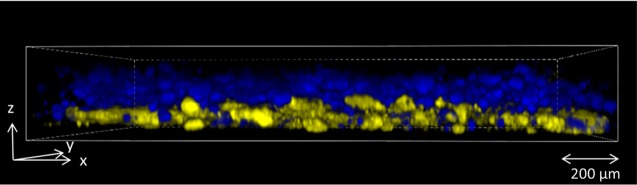
Since some applications may require the printing of multilayered structures containing spatially separated bacterial strains, the internal structure of multilayered printed bacteria was analyzed. Bilayered structures were printed containing engineered Escherichia coli, in which the bacteria in bottom layer of bioink expressed the yellow fluorescent protein mVenus, and the bacteria in the top layer expressed the blue fluorescent protein mCerulean. Each layer was printed using separate tubing and pipet tips to prevent contamination of the top layer by bacteria printed in the previous layer. After 24 h of incubation, the structure was imaged at different depths using confocal microscopy, and the extent of bacterial mixing between the layers was quantified through image analysis. The bottom layer was 81% ± 5% homogeneous, while the top layer was 93% ± 5% homogeneous (Figure 4). This analysis indicates good separation of bacteria between adjacent printed layers, even after extensive periods of incubation. The lower layer may have been less pure due to incomplete solidification before printing of the top layer, which could be improved via an increased waiting time between printing of layers.
Figure 4.
Internal structure of printed layers. Modified strains of E. coli expressing two different fluorescent proteins were printed one on top of the other in a 2-layered square. After 24 h of incubation, the internal structure of the printed bacterial layers was inspected by confocal microscopy. The bottom layer contained 81% ± 5% blue fluorescent cells, while the top layer contained 93% ± 5% yellow cells.
Survival and Metabolic Activity of Printed Bacteria
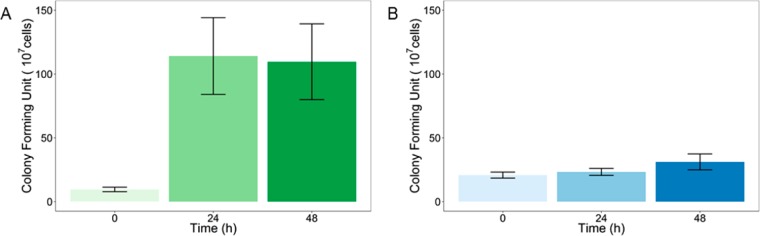
In order for our alignate-based printing system to be successfully applied to microbial materials production, bacteria must be able to survive well within the alginate gel. To test this property, E. coli was incorporated into alginate aerogels, and gels were incubated for varying amounts of time from 0 to 48 h at 37 °C. The gels were then added to a solution of sodium citrate to chelate the calcium ions and dissolve the gel. The samples were grown on LB-agar plates to determine the number of viable cells (colony forming units). An increase in colony forming units observed between the first two data points may indicate that bacterial growth occurs within the alginate gel during the first 24 h after gel production (Figure 5A). Thereafter, colony forming units remained fairly constant for up to 48 h. Comparison with bacteria that were incubated in nonprinted, liquid bioink indicated that the printing process initially reduces the viability of E. coli by approximately 50% (Figure 5B). Thereafter, the levels of viable bacteria in the nonprinted bioink remained nearly constant, likely due to nutrient limitation. The dramatic increase in the number of viable bacteria in the printed gel resulted in an overall increase in viability of approximately 200% in comparison to the nonprinted bioink, which may be due to the additional nutrients in the agar printing substrate and the lower bacterial density after printing. Bacteria can thus remain viable within the alginate gel of our bioink for at least 2 days following gel formation, providing sufficient time for microbial-mediated materials production or patterning to occur.
Figure 5.
Robust bacterial survival within printed alginate gels and planktonic bioink. The number of colony forming units is shown for E. coli printed within alginate gels (A) or as a planktonic sample (B), incubated for varying amounts of time (n = 6). Error bars indicate the standard error.
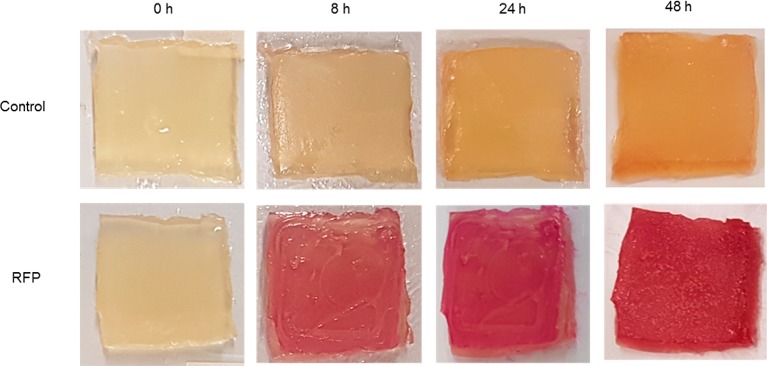
Both survival and metabolic activity of the printed bacteria are key factors to demonstrate the applicability of our printing system. To assess the ability of our printed bacteria to create a product, E. coli containing the rhamnose-inducible red fluorescent protein RFP were printed onto an agar plate containing the rhamnose inducer. The gel was incubated, and the color of the gel was monitored over 48 h. After 8 h of incubation, the induced bioink showed a noticeable red color, which became very intense after 48 h (Figure 6). This experiment demonstrates that our printed bioink is able to support the production of bacterially made materials over short periods of time. Our printing system could be readily applied to the patterned production of bacterially created materials in a variety of different formats. Bioink containing both active bacteria and material precursors could be printed onto a neutral surface, to create a three-dimensionally patterned aerogel within which the bacteria chemically convert the precursors to the desired final product. The thorough commingling of bacteria and chemical substrates within the gel in this configuration would lead to high efficiency of material production. Alternately, alginate gel containing only the chemical precursor could be printed and then immersed within a liquid bacterial culture to create a final 3D-patterned material that is largely bacteria-free. In a third scenario, bacteria-containing bioink could be printed onto a surface that is coated with material precursors, which the bacteria could then convert into a two-dimensionally patterned final product. This approach has the appealing feature that the alginate gel could be dissolved away after the fact, leaving behind only the final material.
Figure 6.
Metabolic activity within printed alginate gels. E. coli with and without a rhamnose-inducible RFP plasmid were printed onto a substrate containing rhamnose. Printed gels were incubated at 37 °C, and color changes were observed over time.
The work shown here demonstrates the development of macroscopic material printing with millimeter-scale resolution using removable aerogels and bacterial chemistry. This approach enables us also to print precursor or supportive material directly with the bacteria. Our printing technique is inexpensive, straightforward, and can produce bacterial structures of a wide variety of three-dimensional shapes without requiring printing scaffolds, excepting structures that contain internal bridges or enclosed hollow spaces. Our technology is well-suited for the use of wild-type organisms or synthetically modified bacteria, which could be designed to carry out new combinations of microbial reactions to create a great number of different types of materials. Connecting our novel and straightforward bacteria printing techniques with approaches of synthetic biology will further improve its value as a “green” material production process and patterning methodology. The ease and simplicity of our printing approach will allow any interested research group to adapt and improve this process at low cost for multiple possible applications.
Materials and Methods
Printing System
The extruder and heater of a standard 3D printer (CoLiDo DIY) were removed and replaced by a pipet tip, a tubing system, and a syringe pump (Figure 1). Silicon tubing (VWR DENE 3100103/25) with an inner diameter of 1 mm and an outer diameter of 3 mm was used to connect a 200 μL pipet tip to a 10 mL syringe. The syringe was loaded with 10 mL of printer bioink and mounted in a syringe pump (ProSense B.V. NE-300). A secondary 200 μL pipet tip was affixed to the printhead (3b in Figure 1A) and connected to silicon tubing to allow for rapid exchange of the pipet tip to deposit a second type of material. Printed objects were drawn in the free online CAD program Tinkercad, sliced via the RepRap Slic3r software, and manually adapted and implemented for printing using CoLiDo software.
Bacterial Strains, Plasmids, and Culturing
Escherichia coli K12 MG1655 was transformed with an SB101 plasmid containing RFP under the control of a rhamnose-inducible promoter. Escherichia coli Top10 cells were transformed with plasmids AM420 or AM421. Cells were cultured overnight in LB media supplemented with the appropriate antibiotic (25 μg/mL ampicillin or kanamycin) at 37 °C with continuous shaking at 250 rpm.
Plasmid AM420 is a p15A-derived plasmid carrying an ampicillin resistance gene, a constitutively expressed lacI gene, and the gene for the blue fluorescent protein mCerulean (gene sequence originally from pZS2–123,29 Addgene plasmid # 26598) behind an IPTG-inducible promoter.
Plasmid AM421 is a pSC101-derived plasmid carrying a kanamycin resistance gene, a constitutively expressed tetR gene, and the gene for the yellow fluorescent protein mVenus (gene sequence originally from mVenus N1,30 Addgene plasmid # 27793) behind an anhydrotetracycline-inducible promoter.
Printer Bioink
To obtain 10 mL of bioink containing E. coli, 10 mL of overnight bacterial culture (OD600 of approximately 2.5) were spun down at 4000 rpm for 3 min and the supernatant discarded. The cells were resuspended in 5 mL of sterile LB medium (Sigma-Aldrich). A 5 mL mixture of sodium alginate (5% w/v, Sigma-Aldrich) was added to the solution, followed by vortexing.
Printing Substrate
A Petri dish (150 mm × 15 mm) was filled with 20 mL of agar (1.5% w/v) dissolved in LB medium. The printing surface was prepared by the equally distributed application of 500 μL 1 M CaCl2 onto the agar surface.
Resolution and Height Measurements
Printing resolution was determined by measuring the width and height of printed samples with a digital caliper (GEDORE No. 711) three times per position for each layer. Prior to the start of printing, the Petri dish lid containing the agar printing surface was affixed to the printer platform using double-faced adhesive tape to prevent changes in the position of the substrate during measurements. Width measurements were recorded for successive layers printed onto the same base layer. Measurement positions within printed ellipses were selected prior to printing: two random points within curved end regions, two random points in straight-edged side regions, and two random points in the transitional area between curved and straight regions. To determine the height of printed samples, the printed structure was removed from the agar printing surface, therefore additional printed layers could not be added post measurement.
Printing and Imaging of Layered Alginate Gels Containing Fluorescent Bacteria
Printing substrate including inducers (1 mM IPTG to induce mCerulean expression, 50 ng/mL anhydrotetracycline to induce mVenus expression) was prepared in a Petri dish. A sheet of dialysis membrane (Spectra/Por 2 Dialysis Tubing, 12–14 kDa MWCO, Spectrum Europe B.V., The Netherlands) was placed on the agar prior to printing, to ensure diffusion of nutrients and inducers from the agar substrate to the printed gel while facilitating eventual transfer of the gel onto a microscope slide. A single rectangular layer of bioink was printed containing E. coli with AM421, then a second rectangular layer of bioink containing E. coli with AM420 was printed on top of the first layer, using separate tubing and pipet tips to avoid bacterial cross-contamination.
After 24 h of incubation at 37 °C, the printed gels and underlying dialysis membranes were sliced with a scalpel. Samples were transferred onto a microscope slide and imaged with a Nikon A1+ fluorescence confocal microscope (magnification 200×, excitation wavelengths 457 or 514 nm, detected wavelengths 465–500 or 525–555 nm for mCerulean or mVenus, respectively).
Bacterial Survival
To determine bacterial survival, 5 mL of an overnight culture of E. coli K12 MG1655 (OD600 of approximately 2.5) was spun down at 4000 rpm for 3 min and the supernatant discarded. The bacterial pellet was resuspended in 5 mL of fresh LB medium containing 2% w/v sodium alginate by vortexing until all ingredients were entirely dissolved. A portion of the bioink was used to print a 6-layered rectangular quadrangle. The gel was allowed to solidify for 30 min before the first sample was taken. The remainder of the bioink was incubated in unsolidified liquid form at 37 °C as a positive control showing planktonic growth. After 0, 24, or 48 h of incubation at 37 °C, 0.3 g of the gel was removed and dissolved in 1.75 mL of 1 M sodium citrate solution. For planktonic samples, 200 μL samples were removed, containing approximately the same volume of bioink as that sampled from the printed gel. Colony forming units (CFU) were determined following the protocol of Karas et al.(31) In short, each sample was serially diluted three times (in 10-fold increments from 10–1 to 10–8), and 5 μL of each dilution was pipetted in triplicate onto a LB-agar plate. The plates were incubated at 37 °C for 24 h, and visible colonies were counted.
Production of RFP by Printed Bacteria
Two 6-layered, square-shaped gels were printed on an LB-agar plate containing 0.2% rhamnose. One gel contained E. coli with a rhamnose-inducible RFP-producing plasmid, and the other one contained wild-type E. coli K12 MG1655. The agar around the printed gel was removed to increase its visibility. The printed gels were photographed under constant light conditions over a 48-h period.
Statistical Methods
R-Studio was used to perform the statistical analyses. All data sets were assumed to be normally distributed and were checked for outliers with a Dixon’s Q-Test. Unless noted, no outliers were removed from the data sets. One-way ANOVA was used to compare multiple data sets, and two-sample Student’s t test was used for comparing two data sets. In cases where the ANOVA test showed a significant difference, a Tukey’s post hoc test was used to obtain an overview of all significant differences occurring within the data set.
Acknowledgments
Our thanks to Roland Kieffer for helping us to develop the 3D printer and to Stan Brouns and Rebecca McKenzie for donating the RFP-producing E. coli strain. The plasmids AM420 and AM421 were constructed and graciously provided by Ferhat Büke. This work was supported by The Netherlands Organization for Scientific Research (NOW/OCW), as part of the Frontiers of Nanoscience program.
Glossary
Abbreviations
- CFU
colony forming unit
- OD
optical density
- TSB
tryptic soy broth
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.6b00395.
Video S1 showing the printer creating a monolayer ellipse in real time (AVI)
The authors declare no competing financial interest.
Supplementary Material
References
- Hiraishi T. (2016) Poly(aspartic acid) (PAA) hydrolases and PAA biodegradation: current knowledge and impact on applications. Appl. Microbiol. Biotechnol. 100, 1623–1630. 10.1007/s00253-015-7216-7. [DOI] [PubMed] [Google Scholar]
- Stanton B. C.; Nielsen A. A.; Tamsir A.; Clancy K.; Peterson T.; Voigt C. A. (2014) Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 10, 99–105. 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J.; Yin P.; Ortiz M. E.; Subsoontorn P.; Endy D. (2013) Amplifying genetic logic gates. Science 340, 599–603. 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- Prindle A.; Selimkhanov J.; Li H.; Razinkov I.; Tsimring L. S.; Hasty J. (2014) Rapid and tunable post-translational coupling of genetic circuits. Nature 508, 387–391. 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden R. A.; Hill D. J.; Kenward M. A.; Williams C. D.; Radecka I. (2007) Bacterial synthesis of biodegradable polyhydroxyalkanoates. J. Appl. Microbiol. 102, 1437–1449. 10.1111/j.1365-2672.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- Sulaeva I.; Henniges U.; Rosenau T.; Potthast A. (2015) Bacterial cellulose as a material for wound treatment: Properties and modifications. A review. Biotechnol. Adv. 33, 1547–1571. 10.1016/j.biotechadv.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Zhong C.; Gurry T.; Cheng A. A.; Downey J.; Deng Z.; Stultz C. M.; Lu T. K. (2014) Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 9, 858–866. 10.1038/nnano.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. Y.; Deng Z.; Billings A. N.; Seker U. O.; Lu M. Y.; Citorik R. J.; Zakeri B.; Lu T. K. (2014) Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523. 10.1038/nmat3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieden D. T.; Meyer A. S.; Aubin-Tam M.-E. (2016) Using bacteria to make improved, nacre-inspired materials. MRS Advances 1, 559–564. 10.1557/adv.2016.150. [DOI] [Google Scholar]
- Hofer P.; Vermette P.; Groleau D. (2011) Introducing a new bioengineered bug: Methylobacterium extorquens tuned as a microbial Bioplastic factory. Bioeng Bugs 2, 71–79. 10.4161/bbug.2.2.15009. [DOI] [PubMed] [Google Scholar]
- Wang G.; Yao L.; Wang W.; Ou J.; Cheng C.-Y.; Ishii H. (2016) xPrint: A Modularized Liquid Printer for Smart Materials Deposition. ACM 5743–5752. 10.1145/2858036.2858281. [DOI] [Google Scholar]
- Matias E.; Rao B. (2015) 3D Printing: On Its Historical Evolution and the Implications for Buisness. Proceedings of PICMET’15 551. 10.1109/PICMET.2015.7273052. [DOI] [Google Scholar]
- Wong K. V.; Hernandez A. (2012) A Review of Additive Manufactoring. ISRN Mech. Eng. 2012, 1. 10.5402/2012/208760. [DOI] [Google Scholar]
- Cooper K. G. (2005) Rapid Prototyping Technology, Marcel Dekker Inc.. [Google Scholar]
- Murphy S. V.; Atala A. (2014) 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785. 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Pati F.; Ha D. H.; Jang J.; Han H. H.; Rhie J. W.; Cho D. W. (2015) Biomimetic 3D tissue printing for soft tissue regeneration. Biomaterials 62, 164–175. 10.1016/j.biomaterials.2015.05.043. [DOI] [PubMed] [Google Scholar]
- Pati F.; Jang J.; Ha D. H.; Won Kim S.; Rhie J. W.; Shim J. H.; Kim D. H.; Cho D. W. (2014) Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 5, 3935. 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher D. (2015) The Top 15 Bioprinters, 3D Printing Industry. [Google Scholar]
- Cui X.; Boland T.; D’Lima D. D.; Lotz M. K. (2012) Thermal inkjet printing in tissue engineering and regenerative medicine. Recent Pat. Drug Delivery Formulation 6, 149–155. 10.2174/187221112800672949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. M.; Stone A. L.; Parkhill R. L.; Stewart R. L.; Simpkins M. W.; Kachurin A. M.; Warren W. L.; Williams S. K. (2004) Three-dimensional bioassembly tool for generating viable tissue-engineered constructs. Tissue Eng. 10, 1566–1576. 10.1089/ten.2004.10.1566. [DOI] [PubMed] [Google Scholar]
- Armstrong J. P.; Burke M.; Carter B. M.; Davis S. A.; Perriman A. W. (2016) 3D Bioprinting Using a Templated Porous Bioink. Adv. Healthcare Mater. 5, 1724–1730. 10.1002/adhm.201600022. [DOI] [PubMed] [Google Scholar]
- Mironov V.; Visconti R. P.; Kasyanov V.; Forgacs G.; Drake C. J.; Markwald R. R. (2009) Organ printing: tissue spheroids as building blocks. Biomaterials 30, 2164–2174. 10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosier G. K. (2011) Methods for Making Construction Material Using Enzyme Producing Bacteria, US patent application US20110262640 A1.
- Connell J. L.; Ritschdorff E. T.; Whiteley M.; Shear J. B. (2013) 3D printing of microscopic bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 110, 18380–18385. 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. K.; Ma P. X. (2001) Ionically crosslinked alginate hydrogels as scaffolds for tissue engineering: part 1. Structure, gelation rate and mechanical properties. Biomaterials 22, 511–521. 10.1016/S0142-9612(00)00201-5. [DOI] [PubMed] [Google Scholar]
- Almqvist K. F.; Wang L.; Wang J.; Baeten D.; Cornelissen M.; Verdonk R.; Veys E. M.; Verbruggen G. (2001) Culture of chondrocytes in alginate surrounded by fibrin gel: characteristics of the cells over a period of eight weeks. Ann. Rheum. Dis. 60, 781–790. 10.1136/ard.60.8.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Yoo Y.; Kim H.; Lee E.; Lee J. Y. (2015) Reduction of graphene oxide/alginate composite hydrogels for enhanced adsorption of hydrophobic compounds. Nanotechnology 26, 405602. 10.1088/0957-4484/26/40/405602. [DOI] [PubMed] [Google Scholar]
- Hernandez D. D. (2015) Factors Affecting Dimensional Precision of Consumer 3D Printing. Int. J. Aviat. Aeronaut. Aerosp. 10.15394/ijaaa.2015.1085. [DOI] [Google Scholar]
- Cox R. S. D. M.; Elowitz M. B. (2010) A synthetic three-color scaffold for monitoring genetic regulation and noise. J. Biol. Eng. 4, 10. 10.1186/1754-1611-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushik S. V. C. H.; Thaler C.; Puhl H. L.; Vogel S. S. (2006) Cerulean, Venus, and VenusY67C FRET reference standards. Biophys. J. 91, L99–L101. 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas V. O.; Westerlaken I.; Meyer A. S. (2015) The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses. J. Bacteriol. 197, 3206–3215. 10.1128/JB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.