Fig. 1.
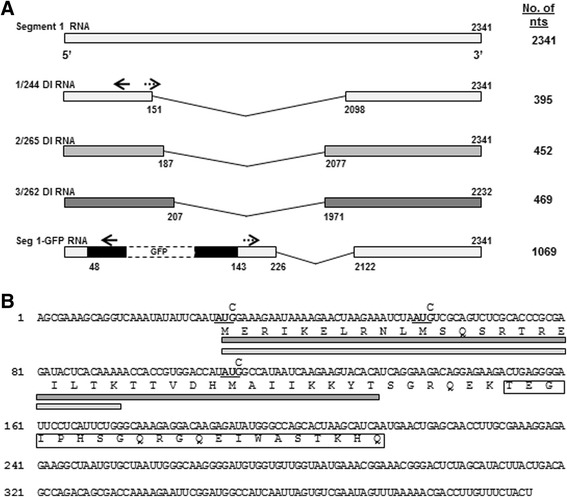
a. Schematic diagram of influenza segment 1244 DI RNA (1/244), segment 2265 DI RNA (2/265), segment 3262 DI RNA (3/262), and the Seg 1-GFP RNA expressed from plasmids. The names derive from the 244, 265 and 262 nt remaining at the 3′ end of the cognate vRNAs respectively.The total number of nucleotides comprising each RNA is shown on the right. The nucleotide positions of the breakpoints that result in the deleted genome RNAs used in this study (positive-sense, 5′ to 3′) are indicated. Solid arrows indicate the primers used in the primer extension assays for cRNA and mRNA analyses and dashed arrows indicate primers used for vRNA analyses. For the construction of Seg 1-GFP RNA see Methods. b. Sequence of 1/244 DI RNA in cRNA sense indicating the open reading frame, and the predicted protein sequence in single letter amino acid code. The 35 residue PB1 binding domain of PB2 is indicated by the dark grey box, and the 22 residue mitochondrial interaction domain of PB2 is indicated by the light grey box. The boxed amino acid sequence downstream from the central deletion that gave rise to 1/244 DI RNA do not arise from the PB2 ORF. The three G → C mutations at nucleotide positions 30, 60 and 111 used to mutate the in-frame AUG initiation codons are shown in bold and underlined
