Abstract
Cancer Stem Cells (CSCs) including Leukemia Stem Cells (LSCs) exhibit self-renewal capacity and differentiation potential, and have the capacity to maintain or renew and propagate a tumor/leukemia. The initial isolation of CSCs/LSCs was in adult myelogenous leukemia, although more recently, the existence of CSCs in a wide variety of other cancers has been demonstrated. CSCs in general, and LSCs specifically in regards to this review, are responsible for initiation of disease, therapeutic resistance and ultimately disease relapse. One key focus in cancer research over the past decade has been to develop therapies to safely eliminate the LSC/CSC population. One major obstacle to this goal is the identification of key mechanisms that distinguish LSCs from normal endogenous hematopoietic stem cells. An additional daunting feature that has recently come to light with advances in next generation sequencing and single cell sequencing is the heterogeneity within leukemias/tumors, with multiple combinations of mutations, gain and loss of function of genes, etc. being capable of driving disease, even within the CSC/LSC population. The focus of this review/perspective will be on our work in identifying and validating in both CML and ALL, a safe and efficacious mechanism to target an evolutionarily conserved signaling nexus, which constitutes a common “Achilles Heel” for LSC/CSC, utilizing small molecule specific CBP/catenin antagonists.
1. Introduction
Stem cells are cells that by definition possess both the capability to self-renew (i.e. give rise to at least one identical daughter cell) as well as differentiate into more mature, specialized cell types. Stem cells can be pluripotent, embryonic stem cells ES or induced pluripotent stem cells (iPS), or of adult tissue origin, termed somatic stem cells (SSC). Somatic stem cells have undergone a partial differentiation process, restricting their differentiation potential, and are hence termed multi-, oligo- or bipotent (1;2). Throughout our lifetime, long-lived, essentially “immortal”, somatic stem cells are called upon to renew and regenerate adult tissues both during homeostatic processes and repair after insult or injury. However, with aging, there is a significant deterioration in stem cell function in a wide array of tissues including blood (lymphoid lineage decreases, myeloid lineage increases and erythroid lineage decreases) (3), which is also associated with increased cancer risk (4). The first type of SSC to be isolated and utilized therapeutically was the hematopoietic stem cell (HSC) in the form of bone marrow for transplantation therapy (5). The dark side of the immortality of SSCs/HSCs is their capacity to be corrupted thereby generating cancer stem cells (CSCs) including leukemia stem cells (LSCs). Like their normal counterparts, CSCs/LSCs exhibit self-renewal capacity and differentiation potential, albeit with aberrant and incomplete differentiation potential, and have the capacity to maintain or renew and propagate a tumor/leukemia. The initial isolation of CSCs/LSCs was in adult myelogenous leukemia (AML)(6), although more recently, the existence of CSCs in a wide variety of other cancers has been demonstrated(7). CSCs in general and specifically in regards to this review, LSCs, are responsible for initiation of disease, therapeutic resistance and ultimately disease relapse (8).
Consequently, one key focus in cancer research over the past decade has been to develop therapies to safely eliminate the CSC/LSC population. A major obstacle to this goal is the identification of key mechanisms that distinguish LSCs from normal endogenous hematopoietic stem cells (HSCs). One additional daunting feature has come to light with recent advances in next generation sequencing and single cell sequencing. It is now clear that cancer is an extremely heterogeneous disease with multiple combinations of mutations, gain and loss of function of genes, etc. being capable of driving disease. Furthermore, within an individual tumor and even within the CSC/LSC population in the tumor, heterogeneity will be a significant problem to overcome (9–11). The focus of this review/perspective will be on our pre-clinical and translational studies in identifying and validating in both CML and ALL, a safe and efficacious mechanism to target the LSC population via a common “Achilles Heel”.
2. Hematopoietic Stem Cells versus Leukemic Stem Cells; More Alike than Different
Unfortunately, from the standpoint of safely targeting LSCs, it appears that the similarities between normal HSCs and LSCs far outweigh the differences between them. (For a recent additional perspective on this topic please see Koeffler and Leong (21)).
This is not all that surprising in that LSCs, in many instances, likely arise from HSCs via mutations (12;13). Importantly, by the definition of “stemness”, they both possess the ability to self-renew and also proceed on to more differentiated cell types. LSCs express similar “stemness” markers and exhibit cellular behaviors highly reminiscent of HSCs. LSCs and HSCs appear to co-inhabit the same specialized niches in the bone marrow and in fact can compete with one another for the limited space within the niche (14–17). Long-lived HSCs are relatively quiescent, infrequently entering cell cycle to maintain homeostasis but more frequently upon injury to repair damaged tissue. Similarly, LSCs appear to be generally quiescent (18). The same signaling pathways involved in regulating LSCs (i.e., Wnt, Notch, Hedeghog, TGFβ/BMP, JAK/Stat, Hippo, MAPK/PI3K) are also involved in the regulation of HSCs (19;20) and multiple points of intersection and crosstalk, including feedback and feedforward loops, connect the various signaling cascades that modulate “stemness”.
3. Wnt Signaling and Stemness
Wnt signaling constitutes an ancient pathway dating back to the early metazoans. The Wnt/catenin pathway is critical throughout normal embryonic development and the life of the organism. It is an extremely complex signal transduction pathway involving 19 mammalian Wnt ligands (22) that trigger a variety of intracellular responses broadly classified as either canonical (increase in nuclear β-catenin) or noncanonical (planar cell polarity, Ca2+/ PKC activation) (23;24). The former is often associated with proliferation and lack of differentiation (for example, as a hallmark of dysregulated Wnt signaling in cancer), whereas the latter is often associated with cell, tissue, and organ differentiation. However, this really is a gross oversimplification (for recent reviews please see (25–27)). β-catenin through its nuclear functions and cytoskeletal/cytoplasmic membrane interactions plays important roles in both canonical and non-canonical Wnt signaling, respectively. In reality, a continuum exists, that coordinates β- catenin-dependent gene expression and cytoplasmic/cytoskeletal β-catenin to affect key developmental and regulatory processes. The entry of β-catenin, or other catenins, for example γ- catenin/plakoglobin (28), into the nucleus and subsequent transcriptional processes are controlled by the so-termed canonical Wnt or Wnt/β-catenin signaling cascade. However, alternative signaling cascades also induce the nuclear translocation of β-catenin and its subsequent participation in transcription. For example, receptor tyrosine kinases (29) and non-receptor tyrosine kinases including Src(30) and Abl(31) can enhance β-catenin-mediated transcription. Additionally, prostaglandins (32), hypoxia (33;34), and high glucose levels (35), also activate Wnt/catenin signaling. These signals are integrated with signals from other key pathways including Notch, JAK/Stat etc., providing nuclear β-catenin with an essential role in balancing self-renewal versus differentiation in adult stem cells (36;37). Wnt signaling is clearly critical in stem cell biology; however, there is no consensus as to whether Wnt signaling is important for either maintenance of potency (8;38) or the differentiation of stem cells (39). Wnt/β-catenin signaling clearly plays dichotomous roles in stem cell biology (38–40).
4. Wnt/Catenin Signaling, Hematopoiesis and HSC, Leukemia and LSC: Differential Coactivator Usage
Wnt signaling, both the canonical and noncanonical pathways, play important roles in hematopoiesis. Retroviral overexpression of activated β-catenin expands the pool of HSCs in long-term cultures and these HSCs activate a LEF-1/TCF reporter in their normal in vivo microenvironment. Inhibitors of the Wnt signaling pathway, as well as ectopic expression of axin or a frizzled ligand-binding domain, results in reduction of HSC growth in vitro and diminished reconstitution in vivo (41).
Genetic deletion of β-catenin during fetal development leads to the impairment of HSC self-renewal. However, several reports have indicated that adult HSCs do not require β-catenin for maintenance (42) and that canonical Wnt signaling regulates hematopoiesis in a dose-dependent fashion (43). Armstrong and coworkers demonstrated that although deletion of β-catenin after CML initiation does not lead to a significant increase in survival, deletion of β-catenin synergizes with imatinib to delay disease recurrence after termination of imatinib treatment. Pharmacologic inhibition of β-catenin using the cyclooxygenase inhibitor indomethacin reduces β-catenin levels and leads to a reduction in LSCs (44). Aberrant Wnt activation need not be cell intrinsic however, as constitutively active β-catenin in osteoblasts is a driver of acute myeloid leukemia (AML) (45). Interestingly, the development of BCR-ABL positive B-ALL and the in vivo self-renewal of B-ALL LSCs were apparently not affected by the absence of β-catenin (46). Furthermore, hematopoiesis occurred normally in the combined absence of β- and γ-catenin using double β/γ-catenin knockout mice (42;47). However, Wnt signaling as judged by Axin2 expression is still maintained and the HSCs maintain long-term repopulation capacity and multilineage differentiation potential, thus pointing to an alternative ‘catenin-like molecule compensating for the loss of both beta and gamma catenin (48).
The Wnt pathway has emerged as a pivotal player in the specification and maintenance of SSC in multiple stem cell niches, in a wide array of tissues and organs including the hematopoietic system (49). It is therefore not surprising that aberrant regulation of Wnt signaling is a recurrent theme in cancer (50;51). This has engendered significant effort to develop therapeutic approaches to target Wnt signaling. A number of factors have thwarted progress in this field, including the enormous complexity of the pathway (23). Further complexity is encountered when targeting transcriptionally active β-catenin, as β-catenin, as well as other catenins (e.g. γ-catenin (52)) can bind a broad spectrum of transcription factors outside of classical Wnt signaling partners, i.e. members of the TCF/LEF family (53). Transcriptionally active β-catenin is associated with an array of biological processes including maintenance of potency, EMT, oxidative stress, and lineage commitment (53). Successful therapeutic manipulation of endogenous “stemness” (normal or cancerous) via modulation of aberrant catenin-regulated transcription offers enormous promise, however it requires significant precision to prevent deleterious effects (e.g., depletion of or increases in somatic mutations) in normal SSC populations (54).
Aberrant Wnt/β- and γ-catenin signaling has been associated with the development of AML (55;56) as well as a critical pathway in the self-renewal of CML LSCs(57;58). Furthermore, transduction of γ-catenin into primitive hematopoietic progenitor cells preserved their immature phenotype during colony formation, suggesting enhanced self-renewal capacity and γ-catenin-transduced cells accelerated the development of leukemia in syngeneic mice (56). However, loss of both β- and γ-catenin leaves Wnt signaling, hematopoiesis and lymphopoiesis intact (59), pointing to yet uncharacterized catenin-like molecule(s) that can compensate for the loss of both β- and γ-catenin.
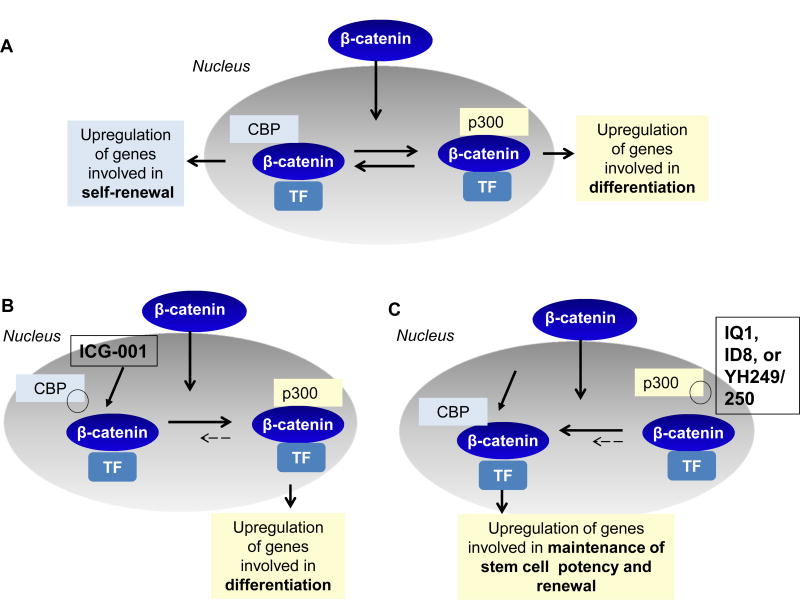
β- (or γ-)catenin recruits one of the two Kat3 transcriptional coactivators, cAMP response element binding protein (CREB-binding protein (CBP) or its closely related homolog, p300 (E1A–binding protein, 300 KDa) as well as other components of the basal transcriptional apparatus to generate a transcriptionally active complex (24;60) (Fig. 1a). The Kat3 coactivator family, CBP and p300, diverged via gene duplication approximately 450 million years ago. These Kat3 coactivators interact with hundreds of proteins in their roles as master orchestrators of transcription. Due to their high degree of protein sequence identity and even higher similarity, they have long been considered largely redundant. However, accumulating evidence has demonstrated that CBP and p300 are not redundant and play definitive and unique roles in vertebrate biology (61–65). Seventeen years ago, from a library of secondary structure mimetics, our lab identified ICG-001 in a forward chemogenomic screen. We subsequently demonstrated that ICG-001 binds specifically and with high affinity (~1 nM) to the N-terminus of CBP (66;67). Over the years, we found that ICG-001, via selectively blocking the CBP/catenin interaction leads to the initiation of differentiation programs in a wide array of stem/progenitor cells (ES, iPS, and SSC) (68;69) (Fig. 1b). Further investigations led to our model of differential coactivator usage, which highlights the distinct roles of the coactivators CBP and p300 in catenin-mediated transcription, particularly within stem/progenitor populations (Fig. 1)(70).Differential utilization of either CBP or p300 as the catenin coactivator is the first decision that guides a stem cell to either maintain potency or initiate a differentiative transcriptional program, respectively (Fig. 1a). We have subsequently identified several small molecules (IQ-1, ID-8, and the specific direct p300/catenin antagonists YH249/250) that selectively antagonize the p300/catenin interaction, thereby enhancing the CBP/catenin interaction, resulting in enhancement of symmetric divisions and the maintenance of potency (pluri- or multipotency) in a variety of stem cell populations (ES, iPS, SSC and HSC) (68;70–73) (Fig. 1c).
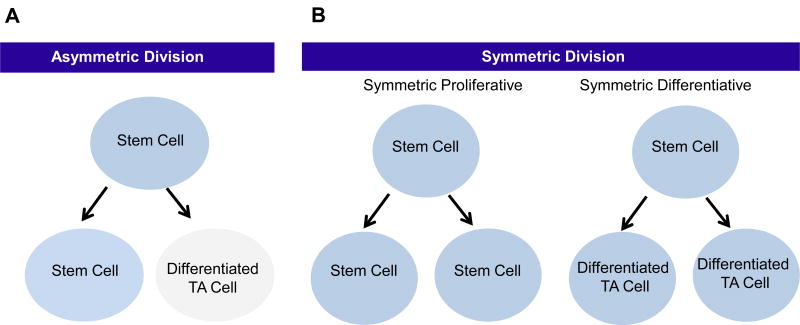
Figure 1.
(A) An asymmetric division results in the production of two daughter cells with different cell fates – one a stem cell and the other a differentiated transient amplifying (TA) cell.
(B) A symmetric proliferative division occurs when the two daughter cells remain as stem cells. A symmetric differentiative division gives rise to two daughter cells, both of which are differentiated TA cells.
5. Wnt Signaling in CML
Despite the stunning clinical success achieved treating chronic-phase (CP) CML patients, responses in advanced-phase patients treated with the TKI Gleevec/Imatinib (IM) are often short-lived, and patients generally undergo disease progression (74). Furthermore, resistance to IM develops in 2–4% of patients annually and IM dose escalation is generally not effective (75).The insensitivity of quiescent CML stem cells to TKIs that express low levels of bcr-abl, has been proposed as a mechanism of resistance(76). Increased nuclear β-catenin has been observed with progression to blast crisis (BC) (77;78). Leukemia stem cells (LSC) are insensitive to TKIs, and additionally, genomic instability in this subpopulation is a significant concern (79). Epigenetic silencing of negative regulators of the Wnt signaling cascade is also frequently observed in leukemias, including CML (80). Chromosomal aberations (81), alterations in the bone marrow microenvironment (82), as well as other mechanisms, may play significant roles in LSC resistance (for a recent review on CML LSC resistance please see T. Holyoake and D. Vetrie (83) ). β-Catenin signaling has also been reported to be activated during the development of MLL (mixed-lineage leukemia) leukemic stem cells(84).
6. Wnt Signaling in ALL
Despite significant progress over the past decades, drug resistance remains a major problem in the treatment of ALL. Dose escalation of current chemotherapeutics is limited by acute and chronic toxicity; therefore new treatment modalities are required. Aberrant Wnt/catenin signaling has been reported to play critical roles in both AML (55) and CML (57;58), where leukemic drug resistant clones have been associated with increased nuclear β-catenin levels (85). However, less is known about the role of Wnt signaling in ALL. Wnt3a has been shown to drive the proliferation of the precursor B-ALL cell lines NALM6, REH and LK63 (86) and endogenous WNT16b expression has been found to be upregulated by the TCF3-PBX1 (E2A–PBX1) fusion (87) . Furthermore, siRNA knockdown of WNT16b, thereby decreasing canonical Wnt/β-catenin signaling, has been shown to initiate apoptosis and reduces the expression of the Wnt-regulated target gene survivin (BIRC5) (87), which has been implicated in both leukemia cell survival and drug resistance (88–90).
7. Wnt signaling in T-ALL
Wnt signaling has been identified as one of the important self-renewal pathways in T-ALL (91;92), Constitutive Wnt/β-catenin signaling was shown to lead to T-ALL in mice (93) . Leukemia stem cells from Ptennull mouse T-ALLs have been shown to have increased levels of β-catenin protein (94) and a real-time, integrated fluorescent Wnt reporter was shown to mark rare leukemia stem cells in T-ALL (95). Over 85% of childhood T-ALL patients showed upregulated β-catenin expression and upregulation of the Wnt target genes axin2, c-myc, tcfl and lef (96). Silencing of β-catenin by small interfering RNA led to increased apoptosis (96). Wnt inhibition by transduction of lentivirus encoding dnTCF into human T-ALL cell lines (HPBALL and RPMI 8402) led to survival prolongation in xenografted T-ALL in mice. Treatment with the tankyrase inhibitor XAV-939 also led to decreased proliferation in vitro (95).
8. Stem Cell Decisions: Symmetry versus Asymmetry
Long-lived HSCs remain relatively quiescent for the majority of their lifetime during normal tissue homeostasis, perhaps dividing only once every few months (97) or even less frequently (98). HSCs can divide either symmetrically or asymmetrically (Fig. 2). Ideally, an asymmetric balance is maintained, whereby one of the daughter cells remains in its niche as a stem cell, while the other daughter proceeds to initiate the differentiation process to maintain tissue homeostasis (Fig. 2a). However, this asymmetric balance is not always maintained and HSCs can also undergo symmetric divisions. There are two modes of symmetric division. In symmetric non-differentiative divisions, both daughter cells remain as stem cells in the niche. Alternatively, HSCs can undergo symmetric differentiative divisions, where both cells leave the niche and go on to differentiate, thereby losing their “stemness” (Fig. 2b). Both modes of symmetric division are presumed to be deleterious to the normal long-lived HSC population, as they can either lead to premature exhaustion of the HSC pool (via symmetric differentiative divisions) or alternatively increase the number of DNA mutations accumulated in the HSC pool (via symmetric non-differentiative divisions). More than 40 years ago, Cairn’s “immortal strand hypothesis” provides a potential, although still controversial, rationale for the preference of HSCs to undergo asymmetric versus symmetric cell divisions (99).
Figure 2.
(A) Upon translocation into the nucleus, β-catenin can associate with the co-factors CBP upregulating ultimately genes involved in self-renewal, or p300 and thereby genes involved in the initiation of differentiation of the CSC/LSC population.
(B) ICG-001 selectively blocks the interaction between β-catenin and CBP. This results in biasing towards p300 usage, and thereby initiates the differentiation transcriptional program with the loss of self-renewal capacity of CSC/LSC.
(C) IQ-1, ID8 (both indirectly) and YH 249/250 (directly) block the interaction between β-catenin and p300. By selectively blocking this interaction, CBP usage is increased, and consequently the initiation of a proliferative or self-renewal transcriptional program is favored.
Although symmetry versus asymmetry is essentially a simple binary decision process, a HSC/LSC located in its niche undergoing mitosis must read an enormously complex array of information from its environment including oxygen levels, nutrient levels, circadian cycles, nervous system innervation, growth factors, adhesion molecules, kinase cascades, cell–cell contacts, etc. to arrive at this eventual binary decision. Interestingly, a bias towards symmetric over asymmetric divisions appears to be a key fundamental difference between LSCs and HSCs. For example, loss of function of the tumor suppressor PTEN leads to premature exhaustion of the normal HSC population (presumably due to increased symmetric differentiative divisions), whereas there is an expansion of the LSC population (presumably due to increased symmetric non-differentiative divisions)(100).More generally, the decision to preferentially undergo symmetric non-differentiative versus symmetric differentiative divisions appears to be an intrinsic difference between cancer stem cells (CSCs) and normal somatic stem cells (SSCs) carrying critical mutations in a number of pathways (i.e., p53, p73, PTEN, Hedgehog, Notch etc.) (101;102). This provides a potential mechanism to attempt to stochastically eliminate mutated defective SSCs prior to the accumulation of additional deleterious mutations (63). A stem cell’s decision to enter cycle and divide symmetrically or asymmetrically or to remain quiescent is clearly governed by the integration of multiple signaling cascades. The key is to understand how this diverse array of signals and crosstalk are integrated and processed into the simple, yet critical decision to divide symmetrically or asymmetrically.
9. Pharmacologically Manipulating HSC and LSC
Over the past 15 years, we have examined the therapeutic potential of selectively antagonizing the CBP/catenin interaction in a variety of preclinical tumor models (both solid and liquid tumors). During the course of our investigations, we observed that CBP/catenin antagonists (i.e. ICG-001), in conjunction with standard chemotherapeutic agents (targeted or cytotoxic agents), demonstrated the ability to safely eliminate drug-resistant CSCs via forced differentiation, without deleterious effects on the normal endogenous stem cell populations and furthermore, ameliorated the toxicity of standard regimens. The differential effects of CBP/catenin antagonists on LSC versus normal HSC and more generally SSC (i.e., forced differentiation and elimination versus differentiation and enhanced repair without depletion) must therefore be cell intrinsic.
CBP/catenin antagonists apparently take advantage of the intrinsic propensity of LSCs to increase the number of symmetric divisions at the expense of asymmetric divisions due to various mutations (e.g., p53, PTEN, etc.) (100;101). Normal long-term repopulating HSCs preferentially divide asymmetrically with one daughter cell remaining in the niche and the other going on to a transient amplifying cell required for hematopoiesis (31), whereas LSCs undergo more symmetric division. We proposed that this fundamental cell intrinsic difference between HSCs and LSCs provides a unique opportunity to therapeutically target and eliminate LSCs without damaging the normal endogenous HSC populations via CBP/catenin antagonists forcing symmetric differentiative divisions in LSCs, while the normal HSCs asymmetrically divide (71;103;104).
10. Targeting CML LSC
The ability to safely eliminate the drug-resistant LSC population, without damaging endogenous HSCs, is critical to the development of more effective chemotherapeutic strategies that completely eliminate the leukemia. We demonstrated that ICG-001, by specifically antagonizing the interaction between CBP and catenin (both β and γ) in CML, initiates a differentiative pathway. This is manifested in the increased expression of myeloid and megakaryocyte differentiation markers including CD11b, CD16, CD33, CD56 and CD41.CBP/catenin antagonists are differentiating agents and not per se cytotoxic, therefore only limited apoptosis is observed. However, the forced differentiation of LSC leads to decreased expression of the antiapoptotic gene survivin, which we have previously shown to be CBP/catenin dependent (61;88), with a concomitant increase in the expression of oncoprotein BCR-ABL. These effects on LSCs are associated with elimination of the quiescent LSC population via forced differentiation into the ‘bulk’ CML population that is sensitive to BCR-ABL antagonists. Downregulation of survivin expression appears to be specific to LSCs (and more generally CSCs). Interestingly, CBP/catenin antagonists do not appear to cause a reduction in survivin expression in normal HSCs or other tissues (epidermal stem cells for example) in vivo based upon the fact that no deleterious effects on these populations have been observed after long term (up to 2 years in mice) administration of CBP/catenin antagonists.
Pretreatment of CML cells in vitro with ICG-001, although not killing the leukemia cells, eliminated, essentially irreversibly, the LSC population, as judged by the lack of engraftment into NSG mice. Importantly, ICG-001 in combination with the second generation TKI Nilotinib, safely eliminated engrafted K562 CML cells as well as primary CML patient samples, in highly immunocompromised NSG mice, without any apparent deleterious effects to the normal endogenous HSC population, as judged by normal hematopoietic parameters and a normal life span (60).
11. Targeting ALL
Sequence or deletion mutations of CBP have recently been identified in ALL (105;106).Extensive analysis of an extended cohort of 71 ALL relapse patients with 270 cases that did not relapse found that 18.3% of relapse cases had sequence or deletion mutations of CBP. In addition, inactivating CBP mutations have been described as a common event in follicular lymphoma and diffuse large B-cell lymphoma (107), the two most frequent forms of B-cell Non-Hodgkin’s lymphoma. Interestingly, of the hundreds of samples sequenced, most mutations occurred within the histone acetyl transferase (HAT) domain with only one described within the N-terminus of CBP, which constitutes both the catenin and ICG-001-binding site. We previously proposed that based upon the critical role for the CBP N-terminal/catenin interaction in maintaining the LSC population, mutations in this N-terminal region would generally not be selected for. Importantly from a therapeutic standpoint, we demonstrated that ICG-001 can also sensitize B-ALL cell lines harboring CBP HAT domain mutations to chemotherapy (104).
The unifying fundamental therapeutic concept therefore, is that in both CML and ALL, CBP/catenin antagonism can deplete drug-resistant LSCs by interruption of self-renewal and shift of catenin/coactivator function (88;90). Antagonizing the CBP/catenin interaction forces symmetric differentiation and additionally down-regulates survivin expression in LSC, thereby sensitizing the cells to chemotherapy, without depletion or deleterious effects on the normal HSCs that undergo asymmetric differentiation. We demonstrated the abrogation of self-renewal by inhibition of serial re-plating of primary ALL cells after treatment with ICG-001. Furthermore, ICG-001 induced, in a dose-dependent manner, the differentiation of murine BCR- ABL1-transformed pre-B cells (CD19+B220+) as determined by analysis of κ-light chain surface expression, a hallmark of B-cell differentiation (104).
12. To the Clinic
In principle, significant concerns about specificity and thereby off-target toxicity, could be raised concerning small molecule inhibitors that target the coactivator protein CBP, as CBP has as many as 500 molecular partners, including a vast array of transcription factors (108). However, these concerns have not been borne out either pre-clinically or even more importantly clinically, utilizing either ICG-001 or the second-generation clinical CBP/catenin antagonist PRI-724(109). This is perhaps at first very surprising. The extremely high biochemical selectivity of ICG- 001/PRI-724 for the N-terminus of its molecular target CBP, the fact that these agents only disrupt a small subset of total CBP interactions and finally the unique evolutionarily conserved roles of the two Kat3 coactivators CBP and p300(63),can be used to rationalize the safety of these agents.
The second-generation specific CBP/catenin antagonistPRI-724 (IC50 ~150 nM) developed by Prism Pharma was safe in preclinical IND enabling toxicology studies, with the no adverse event level being 120 mg/kg/day in dogs given by a 28-day continuous infusion. PRI-724 is in the clinic for both solid tumors (colorectal and pancreatic) and hematopoietic malignancies (CML and AML). An additional trial for HCV induced hepatic fibrosis was also initiated.
An open label, phase Ia safety study in subjects with solid tumors was conducted at USC and reported at ASCO in June 2013 (109). PRI-724 had a very acceptable toxicity profile with dose escalation from 40 to 1280 mg/m2/day with 7 days of continuous i.v. infusion. Downregulation of the biomarker survivin/BIRC5 with upregulation of the differentiation antigen CK20 in CTCs (circulating tumor cells) strongly correlated with increasing plasma concentrations of drug in colorectal cancer patients (109). Additional oncology trials and a trial for HCV-induced hepatic fibrosis with PRI-724 were subsequently initiated (https://clinicaltrials.gov). An open-label, dose-escalation phase Ib/IIa study of PRI-724 for advanced myeloid leukemia at MD Anderson and other cancer centers further demonstrated that PRI-724 is well tolerated. A mechanistic evaluation of patient samples showed that PRI-724 treatment downregulated the expression of the Wnt/catenin targets CD44 and survivin (110).
13. Perspective: CBP/catenin Antagonists: Targeting LSC’s Achilles Heel
As noted in the introduction, tumor heterogeneity and the ability to safely target the drug resistant CSC/LSC population are two common and extremely vexing problems that we must be able to overcome before we can dramatically change overall outcome when treating malignancies. In this respect, CBP/catenin antagonists appear to be truly unique in their ability to target LSCs and more generally drug-resistant CSCs (111;112) in a wide range of tumors that seemingly have little in common with respect to mutational drivers, mechanisms associated with malignancy, i.e. genetic mutations or epigenetic changes, or cell of origin. Even more importantly, CBP/catenin antagonists do so without damaging the normal endogenous stem cell population (71). What provides CBP/catenin antagonists with this unique profile? Quiescence provides long-lived, essentially immortal, HSCs and more generally SSCs, with a safeguard to preserve their functionality by limiting damage to the cell caused by mitochondrial oxidative phosphorylation, DNA damage, and uncontrolled cell cycle entry and exhaustion of the stem cell pool via symmetric differentiative divisions (113;114). HSCs, as well as LSCs, prefer glycolysis rather than oxidative phosphorylation despite the inefficiency in regards to ATP generation (115). The switch from glycolysis to oxidative phosphorylation is associated with activation of quiescent HSCs and the initiation of differentiation (116).
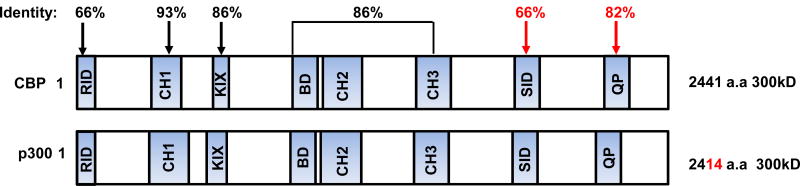
Roughly 450 million years ago, the evolution of vertebrates initiated a new lifestyle requiring critical adaptations, including long-term homeostatic maintenance and tissue repair. This necessitated the advent of SSCs and their corresponding niches, to maintain a relatively quiescent anaerobic metabolic state as opposed to their more proliferative aerobic-differentiated daughter cells, in order to protect the integrity of the genetic material in the stem cell pool (117). This further required a robust, high fidelity mechanism to ensure the proper maintenance of “stemness” in one daughter cell, while in the other daughter cell allowing the initiation of a differentiative program. Intriguingly, the Kat3 coactivator family CBP and p300 diverged via gene duplication apparently just prior to the vertebrate radiation over 450 million years ago. CBP and p300 are extremely large proteins encoded over 33 and 31 exons respectively. Despite having diverged over 450 million years, CBP and p300 retain an extremely high degree of identity, up to 93 %, particularly over a large central core that includes the CH1, KIX, Bromodomain, and CH2 and CH3 regions (Fig. 3)(118;119). Interestingly, the least conserved region, with only 66 % identity between the two Kat3 coactivators, is the extreme N-terminal region, to which both β- (and γ) catenin and the small molecules ICG-001/PRI-724 bind. Yet, the N-terminal regions within each orthologous group are highly conserved for at least the past ~100 million years of evolution; for example, human and mouse CBP are 98 % identical at the amino acid level within this region. One would also assume that over millions of years of evolution that “naturally occurring” CBP/catenin antagonists would have evolved that could assure the asymmetric differentiation of the long lived, highly quiescent SSC pool. Interestingly, the very amino-termini of both CBP and p300 appear to be a nexus for the integration of many signal transduction pathways. For example, a highly conserved LXXLL sequence is present in this region of both CBP and p300, which can recruit nuclear receptor signaling complexes to this region of the Kat3 coactivators. In that sense, we believe that there are numerous naturally occurring CBP/catenin antagonists. For example, all-trans retinoic acid (ATRA) is very effective for the treatment of Acute Promyelocytic Leukemia (APL). Similarly to ICG-001, ATRA does not kill the malignant cells but induces them to differentiate. Vitamin D occupies a prominent position in cancer prevention. Both ATRA and vitamin D, via their respective transcriptional complexes (RAR/RXR and VDR/RXR), can antagonize aberrant Wnt signaling (120) and thereby phenocopy ICG-001/PRI-724, via competition for catenin binding to the N-terminus of CBP.
Figure 3.
Schematic representation of CBP and p300 and the high percentage of identity at the amino acid level between various regions of these largeKAT3 coactivators.
However, synergistic effects on the activation of gene expression by ATRA and Wnt for example (Szeto et al. 2001), have been reported and both vitamin D and ATRA drive the expression of distinct cassettes of genes. We have recently proposed (63) and subsequently have confirmed (Ono et al manuscript submitted) that a highly evolutionarily conserved 27bp deletion in CBP, between the β-catenin-binding region and the LXXLL nuclear receptor binding sequence regulates nuclear receptor antagonism or synergy with Wnt/catenin signaling. ATRA and vitamin D therefore are not simply “pure antagonists” of CBP/catenin signaling and, in that sense, differ from ICG-001/PRI-724. Interestingly, a number of nuclear receptors, both ligand and “orphan” receptors, also demonstrate the ability to either maintain potency or initiate differentiation in stem cell populations, in a similar manner to what we have observed with specific CBP/catenin or p300/catenin antagonists (73;121). For example, PPARδ agonists improved HSC maintenance via increased asymmetric division presumably via CBP/catenin antagonism (122). In that sense, nuclear receptor ligands can behave as CBP/catenin antagonists partially phenocopying ICG-001/PRI-724. However, there are several important differences. Small molecule CBP/catenin antagonists are direct inhibitors (i.e., they bind directly to CBP and do not require any protein cofactors e.g. RAR/RXR) and are pure CBP antagonists (i.e., they have no agonistic activity per se). Furthermore, they allow for stochastic differentiation (i.e., non-deterministic), whereas ATRA or vitamin D, after antagonizing the CBP/catenin interaction, via p300-dependent agonistic properties, bias lineage commitment. We have previously proposed that this p300-dependent lineage biasing is associated with the deleterious effects of high concentrations of ATRA on embryonic development (123), which are not observed in mice treated with ICG-001 in utero (63). Striking differential coactivator usage by the nuclear receptor family has also observed in prostate cancer cells, where 47 % of androgen-regulated genes were p300-dependent, whereas only 0.3 % was CBP-responsive (124). Beyond conserved catenin and nuclear receptor binding regions, the interferon responsive transcription factor Stat1 has also been shown to bind to the very amino terminus of CBP and p300. Further, there are approximately 20 serine and threonine residues that can be post-translationally modified within the first 111 amino acid residues of CBP and p300 (125). In our view, in this fashion the N-termini of the Kat3 coactivators function as a nexus for the integration of an array of signaling cascades that determine the critical stem cell decision to divide symmetrically or asymmetrically, via controlling the balance between the CBP/catenin and the p300/catenin interaction (63). We propose, therefore, that CBP/catenin antagonists can safely target a common “Achilles Heel” in LSC in both CML and ALL and more generally CSCs in other malignancies. CBP/catenin antagonists by specifically and directly binding to CBP initiate differentiation in both LSC and HSC. However, CBP/catenin antagonists take advantage of the preference of LSCs to divide symmetrically relative to asymmetrically, thereby CBP/catenin antagonists via forced differentiation, stochastically “differentiate away” LSC from their niche to more differentiated bulk leukemia and thus targetable by conventional chemotherapy (i.e. Imatinib or cytotoxic agents). However, HSC preferentially divide asymmetrically, thereby always maintaining one stem cell in the niche. Therefore CBP/catenin antagonists do not deplete the endogenous normal HSC pool. Furthermore, by targeting the highly conserved N-terminus of CBP, that appears to be relatively devoid of escape mutations, likely due to its critical role in stem cell maintenance, resistance to CBP/catenin antagonists appears to be quite unusual. Finally, the role of the N-terminus of CBP as a signaling nexus allows CBP/catenin antagonists to work effectively against an enormous range of mutations (p53, PTEN, KRAS, BRAF, APC etc.), signaling networks and epigenetic modifications. The fundamental nature of this balance of coactivator usage by catenin is already manifested at the first cellular decision point in mammalian biology (i.e. at the 8 cell stage of embryogenesis) and we proposed that it is carried through all stem cell populations in vertebrates (63). Thus the ability to target LSCs in ALL and CML, we believe to be just a glimpse of the capacity of CBP/catenin antagonists to safely treat malignancies via elimination of drug-resistant CSCs to provide real cures for these malignancies, similar in this regard to the efficacy of bone marrow transplantation. Further clinical investigation will be needed to confirm this.
Highlights.
Wnt signaling regulates self-renewal and differentiation in normal stem cells.
Aberrant Wnt signaling in leukemia stem cells (LSC) is associated with resistance.
The small molecule ICG-001 selectively blocks the CBP/β- or γ-catenin interaction.
CBP/catenin antagonists may safely target an “Achilles Heel” in LSC.
Acknowledgments
YMK was supported NIH R01CA172896. MK is supported by USC Norris Comprehensive Cancer Center Support Grant P30 CA014089, NIH R01CA166161, R21NS074392, R21AI105057 and NIH R01 HL112638.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
Reference List
- 1.Trentin A, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Self-renewal capacity is a widespread property of various types of neural crest precursor cells. Proc Natl Acad Sci U S A. 2004 Mar 30;101(13):4495–4500. doi: 10.1073/pnas.0400629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013 Jun;140(12):2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Astle CM, Harrison DE. Genetic regulation of primitive hematopoietic stem cell senescence. Exp Hematol. 2000 Apr;28(4):442–450. doi: 10.1016/s0301-472x(99)00157-5. [DOI] [PubMed] [Google Scholar]
- 4.Sharpless NE, Depinho RA. Cancer biology: gone but not forgotten. Nature. 2007 Feb 8;445(7128):606–607. doi: 10.1038/nature05567. [DOI] [PubMed] [Google Scholar]
- 5.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008 Nov 1;112(9):3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997 Jul;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M, Rosen JM. Stem cells in the etiology and treatment of cancer. Curr Opin Genet Dev. 2006 Feb;16(1):60–64. doi: 10.1016/j.gde.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov 1;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Kinstrie R, Copland M. Targeting chronic myeloid leukemia stem cells. Curr Hematol Malig Rep. 2013 Mar;8(1):14–21. doi: 10.1007/s11899-012-0148-8. [DOI] [PubMed] [Google Scholar]
- 10.Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O’Laughlin M, Fronick C, Magrini V, Demeter RT, Fulton RS, Eades WC, Link DC, Graubert TA, Walter MJ, Mardis ER, DiPersio JF, Wilson RK, Ley TJ. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014 Mar 17;25(3):379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, David E, Cohen N, Lauridsen FK, Haas S, Schlitzer A, Mildner A, Ginhoux F, Jung S, Trumpp A, Porse BT, Tanay A, Amit I. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015 Dec 17;163(7):1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013 Jul;27(7):1441–1450. doi: 10.1038/leu.2013.100. [DOI] [PubMed] [Google Scholar]
- 13.Naka K, Hirao A. Maintenance of genomic integrity in hematopoietic stem cells. Int J Hematol. 2011 Apr;93(4):434–439. doi: 10.1007/s12185-011-0793-z. [DOI] [PubMed] [Google Scholar]
- 14.Glait-Santar C, Desmond R, Feng X, Bat T, Chen J, Heuston E, Mizukawa B, Mulloy JC, Bodine DM, Larochelle A, Dunbar CE. Functional Niche Competition Between Normal Hematopoietic Stem and Progenitor Cells and Myeloid Leukemia Cells. Stem Cells. 2015 Dec;33(12):3635–3642. doi: 10.1002/stem.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLean AL, Filippi S, Stumpf MP. The ecology in the hematopoietic stem cell niche determines the clinical outcome in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2014 Mar 11;111(10):3883–3888. doi: 10.1073/pnas.1317072111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Aguilera A, Mendez-Ferrer S. The hematopoietic stem-cell niche in health and leukemia. Cell Mol Life Sci. 2017 Feb;74(4):579–590. doi: 10.1007/s00018-016-2306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shafat MS, Gnaneswaran B, Bowles KM, Rushworth SA. The bone marrow microenvironment - Home of the leukemic blasts. Blood Rev. 2017 Mar 12; doi: 10.1016/j.blre.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Takeishi S, Nakayama KI. To wake up cancer stem cells, or to let them sleep, that is the question. Cancer Sci. 2016 Jul;107(7):875–881. doi: 10.1111/cas.12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaBarge MA. The difficulty of targeting cancer stem cell niches. Clin Cancer Res. 2010 Jun 15;16(12):3121–3129. doi: 10.1158/1078-0432.CCR-09-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010 Jun 15;16(12):3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeffler HP, Leong G. Preleukemia: one name, many meanings. Leukemia. 2017 Mar;31(3):534–542. doi: 10.1038/leu.2016.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013 Jan;41(Database issue):D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012 Dec;13(12):767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 24.Moon RT. Wnt/beta-catenin pathway. Sci STKE. 2005 Feb 15;2005(271) doi: 10.1126/stke.2712005cm1. cm1. [DOI] [PubMed] [Google Scholar]
- 25.Kim YM, Kahn M. The role of the Wnt signaling pathway in cancer stem cells: prospects for drug development. Res Rep Biochem. 2014;4:1–12. doi: 10.2147/RRBC.S53823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birchmeier W. Orchestrating Wnt signalling for metabolic liver zonation. Nat Cell Biol. 2016 Apr 27;18(5):463–465. doi: 10.1038/ncb3349. [DOI] [PubMed] [Google Scholar]
- 27.Staal FJ, Famili F, Garcia PL, Pike-Overzet K. Aberrant Wnt Signaling in Leukemia. Cancers (Basel) 2016 Aug 26;8(9) doi: 10.3390/cancers8090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YM, Ma H, Oehler VG, Gang EJ, Nguyen C, Masiello D, Liu H, Zhao Y, Radich J, Kahn M. The gamma catenin/CBP complex maintains survivin transcription in beta- catenin deficient/depleted cancer cells. Curr Cancer Drug Targets. 2011 Feb;11(2):213–225. doi: 10.2174/156800911794328420. [DOI] [PubMed] [Google Scholar]
- 29.Wagh PK, Gray JK, Zinser GM, Vasiliauskas J, James L, Monga SP, Waltz SE. beta- Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene. 2011 Aug 25;30(34):3694–3704. doi: 10.1038/onc.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coluccia AM, Benati D, Dekhil H, De FA, Lan C, Gambacorti-Passerini C. SKI-606 decreases growth and motility of colorectal cancer cells by preventing pp60(c-Src)- dependent tyrosine phosphorylation of beta-catenin and its nuclear signaling. Cancer Res. 2006 Feb 15;66(4):2279–2286. doi: 10.1158/0008-5472.CAN-05-2057. [DOI] [PubMed] [Google Scholar]
- 31.Ress A, Moelling K. Bcr interferes with beta-catenin-Tcf1 interaction. FEBS Lett. 2006 Feb 20;580(5):1227–1230. doi: 10.1016/j.febslet.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 32.Ishimoto T, Oshima H, Oshima M, Kai K, Torii R, Masuko T, Baba H, Saya H, Nagano O. CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci. 2010 Mar;101(3):673–678. doi: 10.1111/j.1349-7006.2009.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010 Oct;12(10):1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kida A, Kahn M. Hypoxia selects for a quiescent, CML stem/leukemia initiating-like population dependent on CBP/catenin transcription. Curr Mol Pharmacol. 2013 Nov;6(3):204–210. doi: 10.2174/1874467207666140219121219. [DOI] [PubMed] [Google Scholar]
- 35.Chocarro-Calvo A, Garcia-Martinez JM, Ardila-Gonzalez S, De 1 V, Garcia-Jimenez C. Glucose-induced beta-catenin acetylation enhances Wnt signaling in cancer. Mol Cell. 2013 Feb 7;49(3):474–486. doi: 10.1016/j.molcel.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 36.Brembeck FH, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006 Feb;16(1):51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 37.van VW, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, Franken PF, van GL, Meijlink F, van der Valk MA, Kuipers EJ, Fodde R, Smits R. beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011 Sep;60(9):1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004 Jan;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 39.Otero JJ, Fu W, Kan L, Cuadra AE, Kessler JA. Beta-catenin signaling is required for neural differentiation of embryonic stem cells. Development. 2004 Aug;131(15):3545–3557. doi: 10.1242/dev.01218. [DOI] [PubMed] [Google Scholar]
- 40.Hari L, Brault V, Kleber M, Lee HY, Ille F, Leimeroth R, Paratore C, Suter U, Kemler R, Sommer L. Lineage-specific requirements of beta-catenin in neural crest development. J Cell Biol. 2002 Dec 9;159(5):867–880. doi: 10.1083/jcb.200209039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003 May 22;423(6938):409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 42.Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008 Jan 1;111(1):160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- 43.Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, Fibbe WE, van Dongen JJ, Fodde R, Staal FJ. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011 Oct 4;9(4):345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 44.Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW, Armstrong SA. Genetic and pharmacologic inhibition of beta-catenin targets imatinib- resistant leukemia stem cells in CML. Cell Stem Cell. 2012 Apr 6;10(4):412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, Khiabanian H, Lee A, Murty VV, Friedman R, Brum A, Park D, Galili N, Mukherjee S, Teruya-Feldstein J, Raza A, Rabadan R, Berman E, Kousteni S. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014 Feb 13;506(7487):240–244. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007 Dec;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, Huelsken J, Held W. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008 Jan 1;111(1):142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 48.McCrea PD, Gottardi CJ. Beyond beta-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016 Jan;17(1):55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhl SJ, Kuhl M. On the role of Wnt/beta-catenin signaling in stem cells. Biochim Biophys Acta. 2013 Feb;1830(2):2297–2306. doi: 10.1016/j.bbagen.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012 Jun 13;31(12):2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013 Jan;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 52.McCrea PD, Gottardi CJ. Beyond beta-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016 Jan;17(1):55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le NH, Franken P, Fodde R. Tumour-stroma interactions in colorectal cancer: converging on beta-catenin activation and cancer stemness. Br J Cancer. 2008 Jun 17;98(12):1886–1893. doi: 10.1038/sj.bjc.6604401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010 Jun 15;16(12):3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010 Mar 26;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng X, Beissert T, Kukoc-Zivojnov N, Puccetti E, Altschmied J, Strolz C, Boehrer S, Gul H, Schneider O, Ottmann OG, Hoelzer D, Henschler R, Ruthardt M. Gamma- catenin contributes to leukemogenesis induced by AML-associated translocation products by increasing the self-renewal of very primitive progenitor cells. Blood. 2004 May 1;103(9):3535–3543. doi: 10.1182/blood-2003-09-3335. [DOI] [PubMed] [Google Scholar]
- 57.Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007 Dec;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009 Jan;23(1):109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 59.Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008 Jan 1;111(1):160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- 60.Teo JL, Kahn M. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010 Sep 30;62(12):1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 61.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005 May 19;24(22):3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 62.Roth JF, Shikama N, Henzen C, Desbaillets I, Lutz W, Marino S, Wittwer J, Schorle H, Gassmann M, Eckner R. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 2003 Oct 1;22(19):5186–5196. doi: 10.1093/emboj/cdg473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas PD, Kahn M. Kat3 coactivators in somatic stem cells and cancer stem cells: biological roles, evolution, and pharmacologic manipulation. Cell Biol Toxicol. 2016 Feb;32(1):61–81. doi: 10.1007/s10565-016-9318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000 Feb 1;14(3):272–277. [PMC free article] [PubMed] [Google Scholar]
- 65.Yamauchi T, Oike Y, Kamon J, Waki H, Komeda K, Tsuchida A, Date Y, Li MX, Miki H, Akanuma Y, Nagai R, Kimura S, Saheki T, Nakazato M, Naitoh T, Yamamura K, Kadowaki T. Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat Genet. 2002 Feb;30(2):221–226. doi: 10.1038/ng829. [DOI] [PubMed] [Google Scholar]
- 66.McMillan M, Kahn M. Investigating Wnt signaling: a chemogenomic safari. Drug Discov Today. 2005 Nov 1;10(21):1467–1474. doi: 10.1016/S1359-6446(05)03613-5. [DOI] [PubMed] [Google Scholar]
- 67.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Kim HY, Moon SH, Ha JR, Kahn M. A small molecule inhibitor of beta-catenin/CREB- binding protein transcription [corrected] Proc Natl Acad Sci US A. 2004 Aug 24;101(34):12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hasegawa K, Yasuda SY, Teo JL, Nguyen C, McMillan M, Hsieh CL, Suemori H, Nakatsuji N, Yamamoto M, Miyabayashi T, Lutzko C, Pera MF, Kahn M. Wnt signaling orchestration with a small molecule DYRK inhibitor provides long-term xeno-free human pluripotent cell expansion. Stem Cells Transl Med. 2012 Jan;1(1):18–28. doi: 10.5966/sctm.2011-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee ER, Laflamme MA, Papayannopoulou T, Kahn M, Murry CE, Henderson WR., Jr Human embryonic stem cells differentiated to lung lineage-specific cells ameliorate pulmonary fibrosis in a xenograft transplant mouse model. PLoS One. 2012;7(3):e33165. doi: 10.1371/journal.pone.0033165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyabayashi T, Teo JL, Yamamoto M, McMillan M, Nguyen C, Kahn M. Wnt/beta- catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao Y, Masiello D, McMillian M, Nguyen C, Wu Y, Melendez E, Smbatyan G, Kida A, He Y, Teo JL, Kahn M. CBP/catenin antagonist safely eliminates drug-resistant leukemia-initiating cells. Oncogene. 2016 Jul 14;35(28):3705–3717. doi: 10.1038/onc.2015.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008 Aug 7;3(2):132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higuchi Y, Nguyen C, Yasuda SY, McMillan M, Hasegawa K, Kahn M. Specific Direct Small Molecule p300/beta-Catenin Antagonists Maintain Stem Cell Potency. Curr Mol Pharmacol. 2016;9(3):272–279. doi: 10.2174/1874467208666150526155146. [DOI] [PubMed] [Google Scholar]
- 74.Giles FJ, DeAngelo DJ, Baccarani M, Deininger M, Guilhot F, Hughes T, Mauro M, Radich J, Ottmann O, Cortes J. Optimizing outcomes for patients with advanced disease in chronic myelogenous leukemia. Semin Oncol. 2008 Feb;35(1 Suppl 1):S1–S17. doi: 10.1053/j.seminoncol.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 75.Jabbour EJ, Cortes JE, Kantarjian HM. Resistance to tyrosine kinase inhibition therapy for chronic myelogenous leukemia: a clinical perspective and emerging treatment options. Clin Lymphoma Myeloma Leuk. 2013 Oct;13(5):515–529. doi: 10.1016/j.clml.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rice KN, Jamieson CH. Molecular pathways to CML stem cells. Int J Hematol. 2010 Jun;91(5):748–752. doi: 10.1007/s12185-010-0615-8. [DOI] [PubMed] [Google Scholar]
- 77.Jamieson CH, Weissman IL, Passegue E. Chronic versus acute myelogenous leukemia: a question of self-renewal. Cancer Cell. 2004 Dec;6(6):531–533. doi: 10.1016/j.ccr.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 78.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007 Apr;19(2):150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 79.Bolton-Gillespie E, Schemionek M, Klein HU, Flis S, Hoser G, Lange T, Nieborowska-Skorska M, Maier J, Kerstiens L, Koptyra M, Muller MC, Modi H, Stoklosa T, Seferynska I, Bhatia R, Holyoake TL, Koschmieder S, Skorski T. Genomic instability may originate from imatinib-refractory chronic myeloid leukemia stem cells. Blood. 2013 May 16;121(20):4175–4183. doi: 10.1182/blood-2012-11-466938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pehlivan M, Sercan Z, Sercan HO. sFRP1 promoter methylation is associated with persistent Philadelphia chromosome in chronic myeloid leukemia. Leuk Res. 2009 Aug;33(8):1062–1067. doi: 10.1016/j.leukres.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 81.Chen Z, Shao C, Wang W, Zuo Z, Mou X, Hu SJ, DiGiuseppe JA, Zu Y, Medeiros LJ, Hu S. Cytogenetic landscape and impact in blast phase of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Leukemia. 2017 Mar;31(3):585–592. doi: 10.1038/leu.2016.231. [DOI] [PubMed] [Google Scholar]
- 82.Korn C, Mendez-Ferrer S. Myeloid malignancies and the microenvironment. Blood. 2017 Feb 16;129(7):811–822. doi: 10.1182/blood-2016-09-670224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood. 2017 Feb 3; doi: 10.1182/blood-2016-09-696013. [DOI] [PubMed] [Google Scholar]
- 84.Yeung J, Esposito MT, Gandillet A, Zeisig BB, Griessinger E, Bonnet D, So CW. beta- Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010 Dec 14;18(6):606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 85.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, Sawyers CL, Weissman IL. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004 Aug 12;351(7):657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 86.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007 Aug;138(3):338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 87.Mazieres J, You L, He B, Xu Z, Lee AY, Mikami I, McCormick F, Jablons DM. Inhibition of Wnt16 in human acute lymphoblastoid leukemia cells containing the t(1;19) translocation induces apoptosis. Oncogene. 2005 Aug 11;24(34):5396–5400. doi: 10.1038/sj.onc.1208568. [DOI] [PubMed] [Google Scholar]
- 88.Park E, Gang EJ, Hsieh YT, Schaefer P, Chae S, Klemm L, Huantes S, Loh M, Conway EM, Kang ES, Hoe KH, Hofmann WK, Heisterkamp N, Pelus L, Keerthivasan G, Crispino J, Kahn M, Muschen M, Kim YM. Targeting survivin overcomes drug resistance in acute lymphoblastic leukemia. Blood. 2011 Aug 25;118(8):2191–2199. doi: 10.1182/blood-2011-04-351239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tyner JW, Jemal AM, Thayer M, Druker BJ, Chang BH. Targeting survivin and p53 in pediatric acute lymphoblastic leukemia. Leukemia. 2012 Apr;26(4):623–632. doi: 10.1038/leu.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morrison DJ, Hogan LE, Condos G, Bhatla T, Germino N, Moskowitz NP, Lee L, Bhojwani D, Horton TM, Belitskaya-Levy I, Greenberger LM, Horak ID, Grupp SA, Teachey DT, Raetz EA, Carroll WL. Endogenous knockdown of survivin improves chemotherapeutic response in ALL models. Leukemia. 2012 Feb;26(2):271–279. doi: 10.1038/leu.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weerkamp F, van Dongen JJ, Staal FJ. Notch and Wnt signaling in T-lymphocyte development and acute lymphoblastic leukemia. Leukemia. 2006 Jul;20(7):1197–1205. doi: 10.1038/sj.leu.2404255. [DOI] [PubMed] [Google Scholar]
- 92.Belmonte M, Hoofd C, Weng AP, Giambra V. Targeting leukemia stem cells: which pathways drive self-renewal activity in T-cell acute lymphoblastic leukemia? Curr Oncol. 2016 Feb;23(1):34–41. doi: 10.3747/co.23.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo Z, Dose M, Kovalovsky D, Chang R, O’Neil J, Look AT, von BH, Khazaie K, Gounari F. Beta-catenin stabilization stalls the transition from double-positive to singlepositive stage and predisposes thymocytes to malignant transformation. Blood. 2007 Jun 15;109(12):5463–5472. doi: 10.1182/blood-2006-11-059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo W, Lasky JL, Chang CJ, Mosessian S, Lewis X, Xiao Y, Yeh JE, Chen JY, Iruela-Arispe ML, Varella-Garcia M, Wu H. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008 May 22;453(7194):529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Giambra V, Jenkins CE, Lam SH, Hoofd C, Belmonte M, Wang X, Gusscott S, Gracias D, Weng AP. Leukemia stem cells in T-ALL require active Hif1alpha and Wnt signaling. Blood. 2015 Jun 18;125(25):3917–3927. doi: 10.1182/blood-2014-10-609370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ng OH, Erbilgin Y, Firtina S, Celkan T, Karakas Z, Aydogan G, Turkkan E, Yildirmak Y, Timur C, Zengin E, van Dongen JJ, Staal FJ, Ozbek U, Sayitoglu M. Deregulated WNT signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J. 2014 Mar 14;4:e192. doi: 10.1038/bcj.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Foudi A, Hochedlinger K, Van BD, Schindler JW, Jaenisch R, Carey V, Hock H. Analysis of histone 2B–GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009 Jan;27(1):84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baker AM, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, Humphries A, Elia G, McDonald SA, Wright NA, Simons BD, Jansen M, Graham TA. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014 Aug 21;8(4):940–947. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975 May 15;255(5505):197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 100.Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010 Nov 5;7(5):593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self- renewing divisions in mammary stem cells. Cell. 2009 Sep 18;138(6):1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 102.Ferent J, Cochard L, Faure H, Taddei M, Hahn H, Ruat M, Traiffort E. Genetic activation of Hedgehog signaling unbalances the rate of neural stem cell renewal by increasing symmetric divisions. Stem Cell Reports. 2014 Aug 12;3(2):312–323. doi: 10.1016/j.stemcr.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Teo JL, Ma H, Nguyen C, Lam C, Kahn M. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A. 2005 Aug 23;102(34):12171–12176. doi: 10.1073/pnas.0504600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gang EJ, Hsieh YT, Pham J, Zhao Y, Nguyen C, Huantes S, Park E, Naing K, Klemm L, Swaminathan S, Conway EM, Pelus LM, Crispino J, Mullighan CG, McMillan M, Muschen M, Kahn M, Kim YM. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene. 2014 Apr 24;33(17):2169–2178. doi: 10.1038/onc.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, Buetow KH, Pui CH, Baker SD, Brindle PK, Downing JR. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011 Mar 10;471(7337):235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, Gerhard DS, Loh ML, Reaman GH, Relling MV, Camitta BM, Bowman WP, Smith MA, Willman CL, Downing JR, Hunger SP. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011 Sep 15;118(11):3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, Chan J, Bhagat G, Chadburn A, Gaidano G, Mullighan CG, Rabadan R, Dalla-Favera R. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011 Jul 31;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ring A, Kim YM, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 2014 Aug;10(4):512–525. doi: 10.1007/s12015-014-9515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.El-Khoueiry ABNYYDCSKMZMBJFMITKHLH. A phase I first-in-human study of PRI-724 in patients (pts) with advanced solid tumors. J Clin Oncol. 2013;31(suppl) abstract 2501, Ref Type: Abstract. [Google Scholar]
- 110.Zhou HS, Carter BZ, Andreeff M. Bone marrow niche-mediated survival of leukemia stem cells in acute myeloid leukemia: Yin and Yang. Cancer Biol Med. 2016 Jun;13(2):248–259. doi: 10.20892/j.issn.2095-3941.2016.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wend P, Fang L, Zhu Q, Schipper JH, Loddenkemper C, Kosel F, Brinkmann V, Eckert K, Hindersin S, Holland JD, Lehr S, Kahn M, Ziebold U, Birchmeier W. Wnt/beta- catenin signalling induces MLL to create epigenetic changes in salivary gland tumours. EMBO J. 2013 Jul 17;32(14):1977–1989. doi: 10.1038/emboj.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chan KC, Chan LS, Ip JC, Lo C, Yip TT, Ngan RK, Wong RN, Lo KW, Ng WT, Lee AW, Tsao GS, Kahn M, Lung ML, Mak NK. Therapeutic targeting of CBP/beta- catenin signaling reduces cancer stem-like population and synergistically suppresses growth of EBV-positive nasopharyngeal carcinoma cells with cisplatin. Sci Rep. 2015 Apr 21;5:9979. doi: 10.1038/srep09979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bakker ST, Passegue E. Resilient and resourceful: genome maintenance strategies in hematopoietic stem cells. Exp Hematol. 2013 Nov;41(11):915–923. doi: 10.1016/j.exphem.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008 Feb;9(2):115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 115.Kohli L, Passegue E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends Cell Biol. 2014 Aug;24(8):479–487. doi: 10.1016/j.tcb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Panopoulos AD, Izpisua Belmonte JC. Anaerobicizing into pluripotency. Cell Metab. 2011 Aug 3;14(2):143–144. doi: 10.1016/j.cmet.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 117.Trosko JE, Kang KS. Evolution of energy metabolism, stem cells and cancer stem cells: how the warburg and barker hypotheses might be linked. Int J Stem Cells. 2012 May;5(1):39–56. doi: 10.15283/ijsc.2012.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A–associated p300 and CREB- associated CBP belong to a conserved family of coactivators. Cell. 1994 Jun 17;77(6):799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 119.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A–associated 300- kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994 Apr 15;8(8):869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 120.Dillard AC, Lane MA. Retinol decreases beta-catenin protein levels in retinoic acid- resistant colon cancer cell lines. Mol Carcinog. 2007 Apr;46(4):315–329. doi: 10.1002/mc.20280. [DOI] [PubMed] [Google Scholar]
- 121.Mullen EM, Gu P, Cooney AJ. Nuclear Receptors in Regulation of Mouse ES Cell Pluripotency and Differentiation. PPAR Res. 2007;2007:61563. doi: 10.1155/2007/61563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012 Sep;18(9):1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009 Oct 15;122(Pt 20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ianculescu I, Wu DY, Siegmund KD, Stallcup MR. Selective roles for cAMP response element-binding protein binding protein and p300 protein as coregulators for androgen- regulated gene expression in advanced prostate cancer cells. J Biol Chem. 2012 Feb 3;287(6):4000–4013. doi: 10.1074/jbc.M111.300194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci U S A. 1996 Dec 24;93(26):15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]