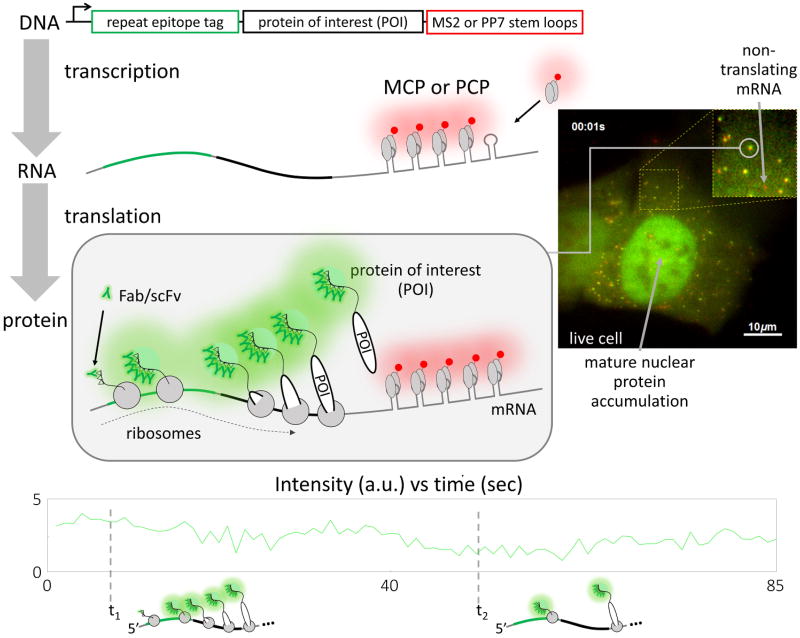
Figure 3. Imaging gene regulatory dynamics in live cells using antibody-based probes.
To successfully image live cell translation dynamics, 3′UTR stem loops and a repeat epitope tag need to be encoded into the gene of interest. During transcription, the stem loops will form (gray loops) and quickly be bound by a coat protein (MS2 or PP7; gray ovals) fused to a fluorescent protein tag (red circle). This enables the visualization of mature mRNA molecules in living cells. When these marked mRNA are translated, the repeat epitopes (triangles) will emerge from the ribosome and be quickly bound by a fluorescent probe (Fab or scFv; glowing “Y”). This will quickly bring light to the growing nascent chain of the protein of interest (POI) while it is being translated. The co-migration of the red mRNA signal and the green nascent chain signal will mark sites of translation. A sample live-cell image is shown, where the green signal corresponds to Fab labeling spaghetti monster tagged lysine demethylase (KDM5B) and the red signal corresponds to MS2-labeled mRNA. These signals colocalize to form yellow spots that mark the sites of translation. The green fluorescence time series from the circled yellow spot in the inset is shown below, along with cartoons depicting how ribosomes might be distributed along the mRNA at the two specified time points. A full movie of this cell can be seen in Supplementary Movie 1. Scale bars, 10 μm.