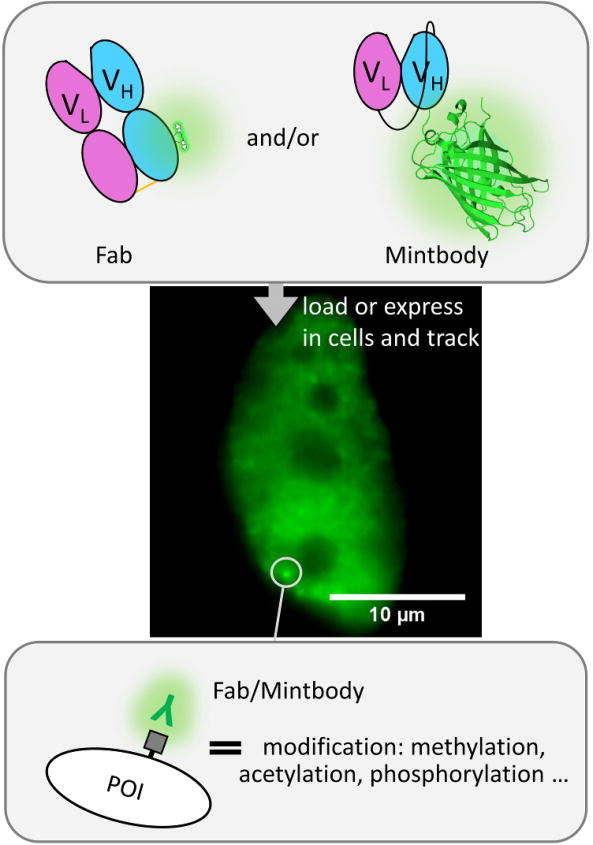
Figure 4. Marking endogenous modifications with antibody-based probes.
By loading fluorescently conjugated Fab (green and pink ovals with small glowing molecule) and/or an scFv-GFP (pink and blue ovals with glowing beta-barrel structure; PDBID: 4KW4), modified proteins can be dynamically labeled in living cells. In this example a Fab/Mintbody (glowing “Y”) is targeting residue-specific modifications (gray square) of an endogenous protein of interest (POI). The modifications accumulate in the nucleus, so Fab/Mintbody accumulate there as well. The fluctuations in the accumulations of the Fab/Mintbody in specific sub-nuclear regions can be used to quantify modification dynamics. The sample cell shown shows the accumulation of Fab (green) labeling endogenous histone H3 Lysine 27 acetylation. Scale bar, 10 μm.