Figure 6.
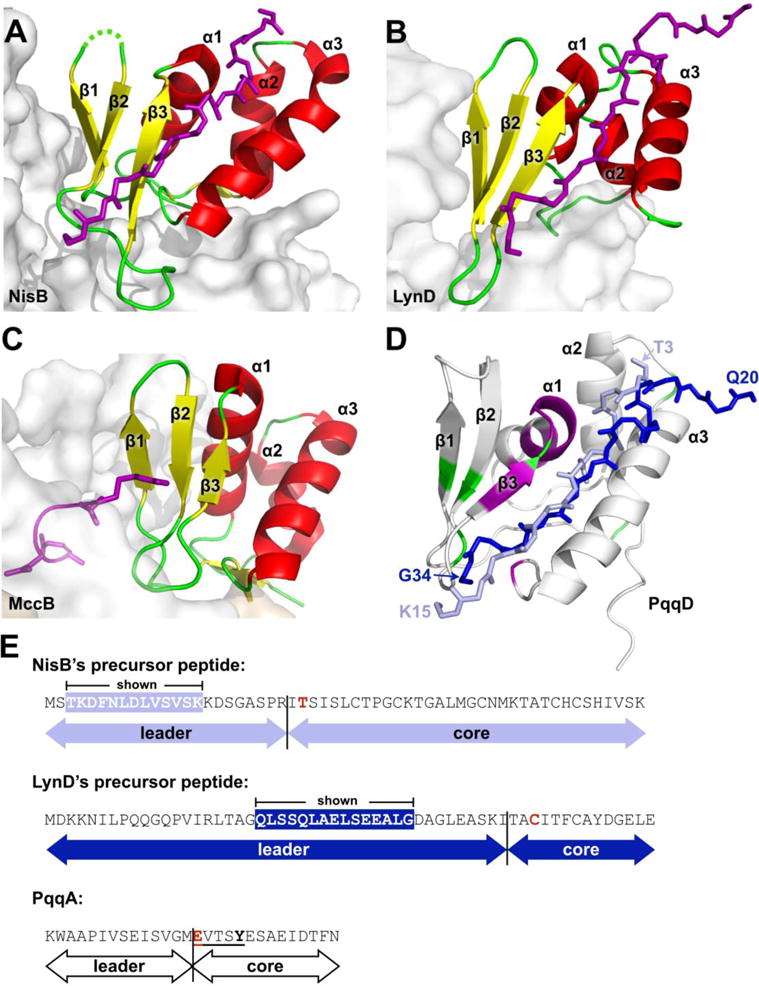
Comparison of PqqD to RREs from other PRPS enzymes in complex with peptide. (A) NisB, PDB 4WD9 (residues 142–223), (B) LynD, PDB 4V1T (residues 1–81), and (C) MccB, PDB 3H9J (residues 1–78). Constituent RREs with bound peptides are shown as cartoon and colored by secondary structure with bound precursor peptides drawn as stick and colored purple. The enzyme of which the RRE is a part is shown as a gray molecular surface. (D) PqqD residues perturbed by binding PqqA and the rSAM enzyme, PqqE, are colored purple and green, respectively. The peptides of NisB and LynD crystal structures are overlaid based on superposition of the RREs with PqqD (NisB peptide, light blue; LynD peptide, dark blue). (E) Precursor peptide sequence comparison between NisB, LynD, and PqqD. The peptide residues observed in the NisB and LynD crystal structures are white in colored boxes. The most proximal residue that is post-translationally modified is colored red.
