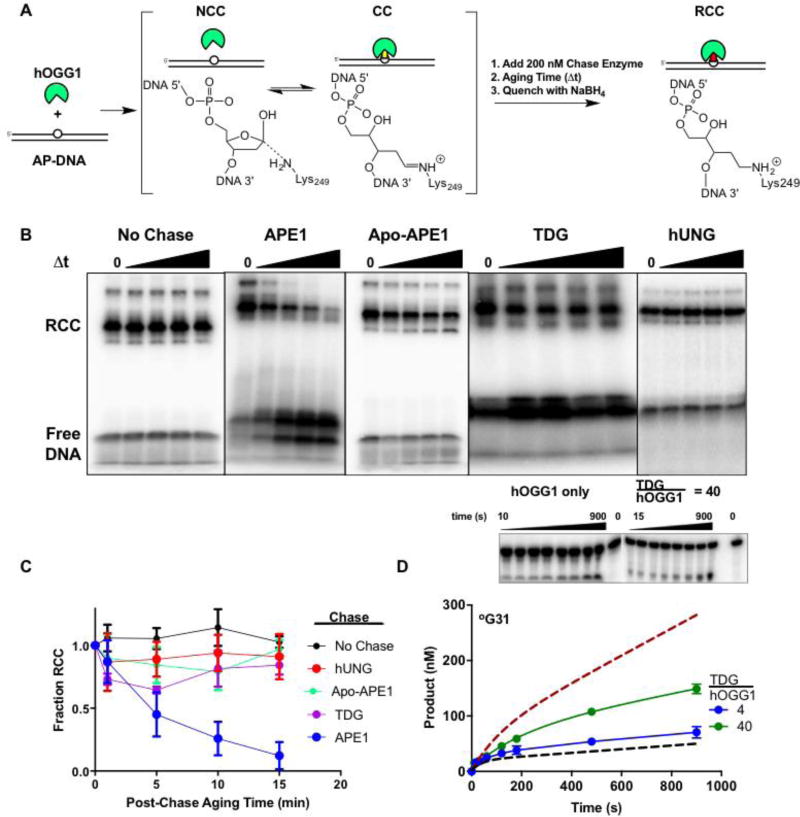
Figure 5. Effects of AP site binding enzymes on dissociation of hOGG1 from an AP site.
(A) The experimental strategy was to incubate hOGG1 with AP-DNA until a reversible equilibrium was established between the covalent Schiff base complex (CC) and the noncovalent complex (NCC). A chase enzyme (APE1, TDG, hUNG, or no enzyme) was then added and after an aging time, NaBH4 was added to reduce the imino linkage between the DNA and Lys249 of hOGG1 (RCC). The chase enzymes were each present at a 200 nM concentration. (B) The reaction products as a function of aging time. The RCC and free DNA species were resolved by SDS-PAGE. Representative gel images for no chase and each chase enzyme are shown. (C) Disappearance of the RCC over 15 minutes using various chase enzymes. (D) TDG modestly stimulates hOGG1 turnover. A 4-fold molar excess of TDG over hOGG1 was not significantly stimulatory (blue), while a 40-fold molar excess (1000 nM) increased the turnover rate by about 2.5 fold (green). The dashed lines are the theoretical curves for reactions with hOGG1 only and in the presence of one equivalent of APE1 (see Fig. 2A). The concentrations of °G31 substrate and hOGG1 were 500 and 25 nM respectively. Experiments were carried out in triplicates and standard errors were plotted.