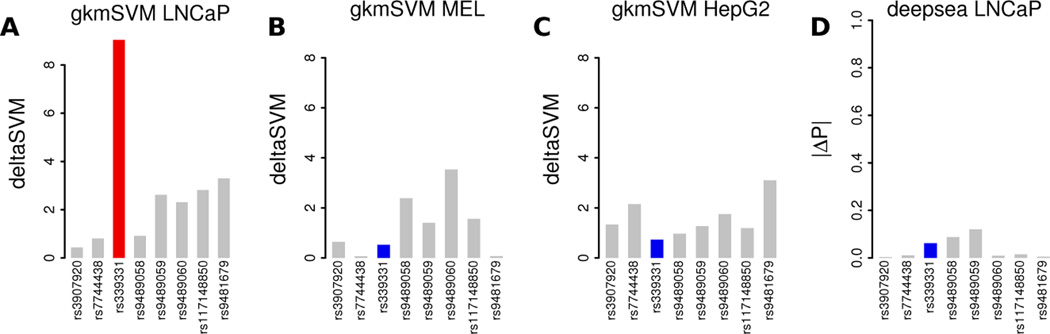
Fig. 5. Predicting causal SNPs within the RFX6 prostate cancer locus.
deltaSVM using a gkm-SVM trained on a prostate cancer cell line LNCaP (A) can identify the causal SNP (red) from among flanking SNPs (grey), but a gkm-SVM trained on melanocytes (B) or HepG2 (C) cannot. Deepsea predictions include LNCaP cells in the training set but do not correctly identify the validated SNP (D).