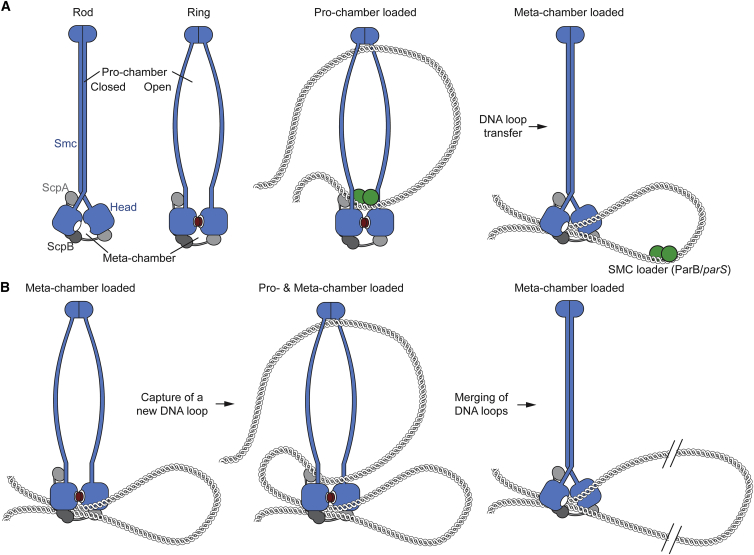
Figure 7.
A Tentative Model for DNA Loop Extrusion by a Double-Chamber SMC Complex
(A) Chromosomal loading. The head-engaged open form of Smc-ScpAB initiates chromosomal DNA transactions by capturing a DNA loop in its DNA pro-chamber, i.e., the Smc interarm space. The loading machinery (in green colors) provides specificity by binding to the open Smc arms. ATP hydrolysis destabilizes the open Smc ring and triggers re-formation of a Smc rod starting from the Smc hinge. Rod formation closes the pro-chamber and pushes DNA into the meta-chamber, i.e., the inter-head/kleisin/kite space.
(B) Processive DNA loop extrusion. After successful loading, the subsequent loop capture-merging cycles start with the engagement of the Smc heads to open up the pro-chamber for DNA loading. To drive directional DNA extrusion, the newly captured DNA must be derived from DNA flanking the already captured DNA loop rather than the DNA loop itself. The asymmetry of Smc-ScpAB might ensure the directionality of this step. Upon ATP hydrolysis, the newly captured DNA is then merged with the DNA loop previously loaded into the meta-chamber to generate a larger DNA loop. A related model for DNA translocation of SMC double-ring “handcuff” complexes is shown in Figure S7.
See also Figure S7.