Figure 2.
Large Heterochromatin Domains Form upon Removal of mst2+
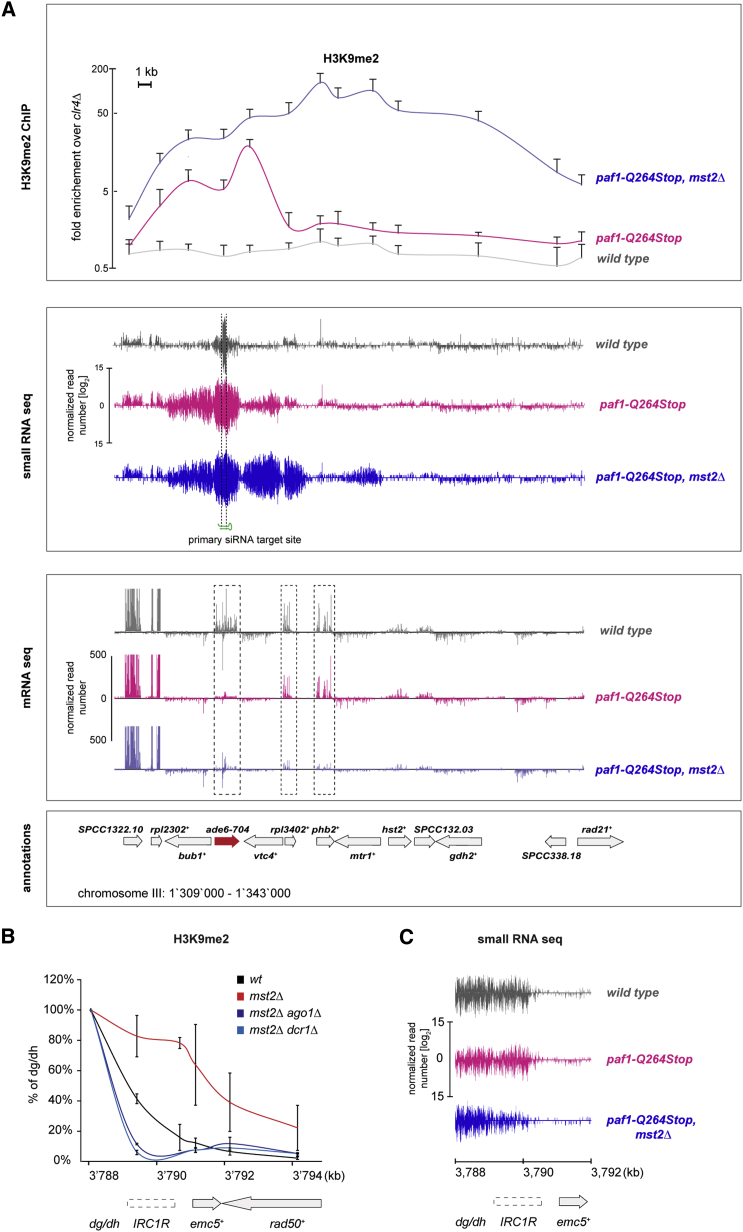
(A) Upper panel: ChIP analysis of H3K9me2 showing enrichments at the target gene ade6-704 and neighboring regions. Error bars indicate SD (n ≥ 3 independent biological replicates). The y axis is shown in logarithmic scale. Middle and lower panels: siRNAs (middle panel) and RNA (lower panel) reads mapping to the ade6-M210 locus and neighboring regions in wild-type (gray), paf1-Q264Stop (red), and paf1-Q264Stop mst2Δ cells (blue), respectively, are shown. Read counts were normalized to the total read number and are depicted in log2 (middle panel) or linear scale (lower panel).
(B) H3K9me2 enrichments at the right centromere boundary of chromosome 1 (IRC1R). ChIP enrichments are shown relative to the centromeric repeats dg/dh, which was set at 100%. Error bars indicate SD (n = 2 or 3 independent biological replicates; mst2Δ ago1Δ and mst2Δ dcr1Δ or wild-type (WT) and mst2Δ, respectively).
(C) siRNAs mapping to IRC1R and neighboring regions in wild-type (gray), paf1-Q264Stop (red), and paf1-Q264Stop mst2Δ cells (blue). Read counts were normalized to the total read number and are depicted in log2 scale.
See also Figure S1.