Figure 6.
Mst2-Mediated Acetylation of Brl1 Represses Initiation of Heterochromatin Assembly
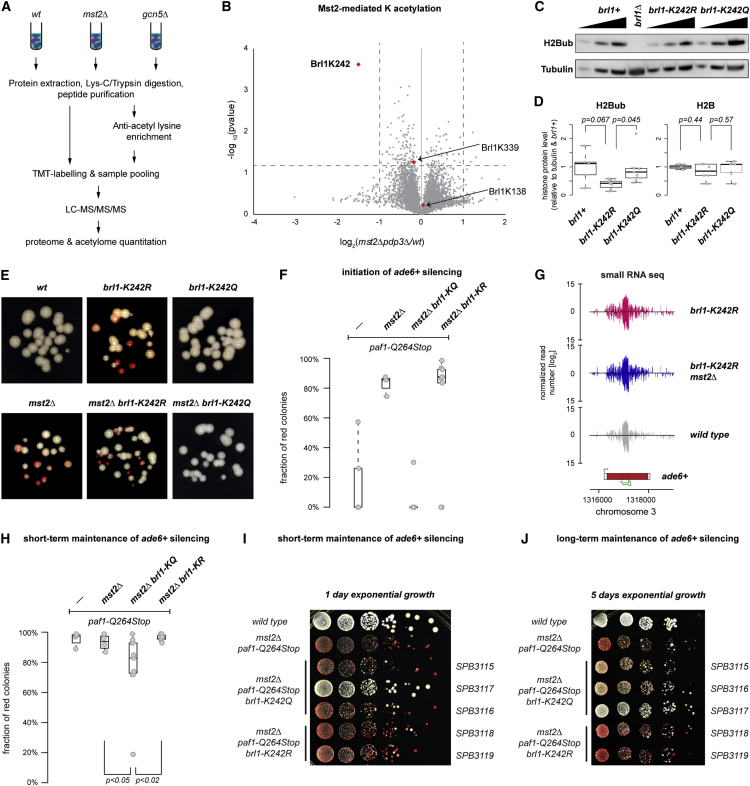
(A) Scheme: acetylomics workflow. Total peptides or peptides enriched for acetylation were labeled with TMT and subjected to LC-MS/MS/MS. We identified 8,926 acetylated peptides and quantified 3,933 proteins (Table S1). See the STAR Methods for more information.
(B) Volcano plot showing fold changes in pdp3Δ mst2Δ compared to WT cells. Identified acetylated Brl1 peptides are shown in red (n = 3 independent biological replicates). The x axis is shown in log2 scale.
(C) Immunodetection of H2BK119ub in different strains. Dilution series was 1/9, 1/3, and 1/1 of the respective protein extracts. Tubulin served as a loading control. A representative experiment is shown.
(D) Quantification of H2BK119ub (left) and H2B (right) levels normalized to tubulin and relative to WT (brl1+). Multiple independent biological replicates were for H2BK119ub (WT n = 5 and brl1-KR/KQ n = 7) and H2B (WT n = 3 and brl1-KR/KQ n = 6). The p values were calculated using the two-sided, two-sample Student’s t test with equal/unequal variance according to prior evaluation with the F test.
(E) Silencing assays of siRNA-directed de novo heterochromatin assembly (as described in Figure 1) in the strains indicated (close up). All strains contained a paf1+ allele. A representative experiment is shown.
(F and H) Initiation (F) and maintenance (H) frequencies in paf1-Q264Stop cells with additional mutations in mst2+ and brl1+. Assessment of initiation/maintenance frequency was as in Figure 1D. The p values were calculated using two-sided, two-sample Student’s t test (n ≥ 8 individual white colonies). Exact numbers are listed in the STAR Methods.
(G) siRNA reads mapping to the ade6-M210 locus and neighboring regions in brl1-K242R (red) and mst2Δbrl1-K242R (blue) cells. Read counts were normalized to total read number and are depicted in log2 scale.
(I and J) Dilution assays showing gradual loss of ectopic silencing at the trp1+::ade6+ locus in brl1-K242Q mutants, but not in brl1-K242R mutants. Cells were grown exponentially for 1 day (I) or 5 days (J), and equal cell numbers were plated onto yeast extract-nourseothricin (YE-Nat). A representative experiment with independent strains is shown. Different yeast strains are depicted on the right.
See also Figures S3 and S4 and Table S1.