Figure 2.
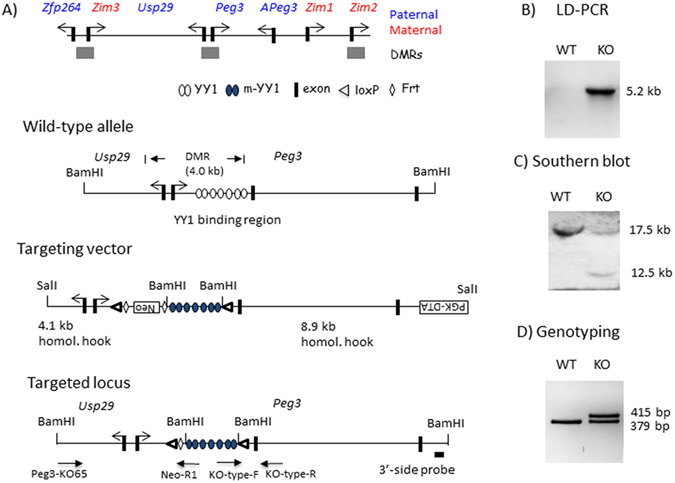
Peg3 domain and the targeting scheme. (A) Schematic representations of the Peg3 domain (upper panel). Each imprinted gene is indicated with an arrow. The paternally and maternally expressed genes are indicated with blue and red, respectively. The three DMRs are indicated with gray boxes. Targeting scheme (lower panel). The 4.0-kb Peg3-DMR contains the first exons of Peg3 and Usp29 and the 2.5-kb YY1 binding region. The transcriptional direction of Peg3 and Usp29 is indicated with arrows, and exons are indicated with thick vertical lines. The region corresponding to the neomycin resistance gene (NeoR) along with the two flanking FRT sites within the targeting vector are indicated by an open box and diamonds, respectively. Arrows underneath ‘Targeted locus’ indicate primers with relative positions that were used for long-distance PCR and genotyping. (B) LD (Long Distance)-PCR. A set of primers, KO65 and NeoR1, successfully amplified the 5.2-kb genomic fragment from the genomic DNA of a heterozygote animal (KO), but not from that of a wild-type littermate (WT), confirming the proper targeting of the KO construct. (C) Southern blot analyses on the DNA isolated from ES cells that had been transfected with the targeting vector. The BamHI-digested DNA hybridized with the 3′side probe shows the wild-type allele (17.5 kb) and the targeted allele (12.5 kb). (D) PCR-based genotyping. The PCR scheme targeting the 2nd loxP site with two primers (KO-type-F and -R) derived two DNA fragments representing the wild-type (379 bp) and KO (415 bp) alleles in the heterozygotes.
