Figure 1.
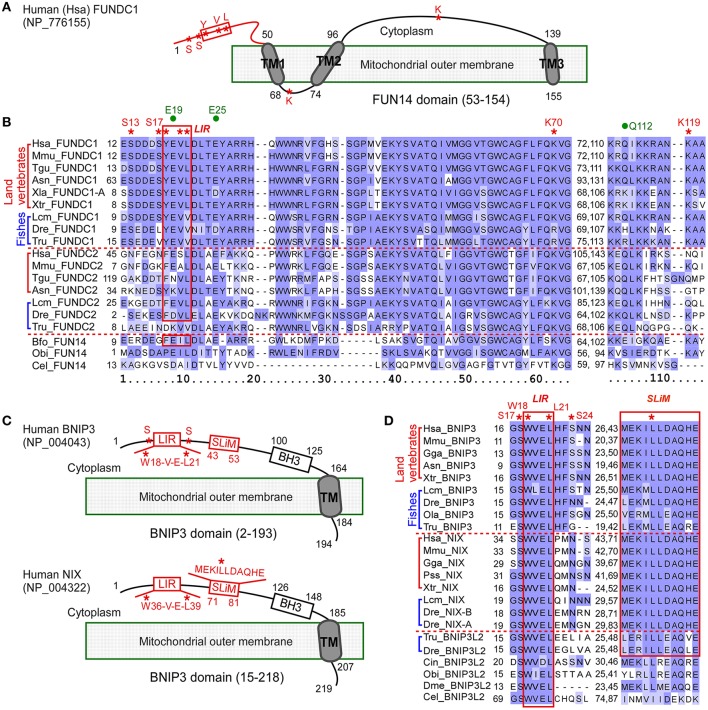
Multiple sequence alignment of proteins containing FUN14 or BNIP3 domain in animals. (A) Illustration representing the topology of Homo sapiens FUNDC1 (NP_776155), a mitochondrial outer membrane protein. Cytosolic region is at the top of the membrane. FUN14 domain (PF04930) covers all three transmembrane (TM) domains. Red box indicates LC3 binding region (LIR, Y/W/F-x-x-L/I/V). Red asterisks indicate key phosphorylation sites (S13, S17, and Y18), conserved sites in LIR, and key sites (K70, K119) for interacting with OPA1 and MARCH5. (B) Alignment of FUN14 domain-containing proteins in representative metazoan species of six land vertebrates, three fishes and three invertebrates. The LIR motif is enclosed within a red box. Red asterisks indicate biologically important residues as labeled in (A). Three green dots indicate predicted functionally divergent sites (see Materials and Methods). (C) Topology of two other mitochondrial outer membrane proteins, BNIP3 (NP_004043) and its homolog NIX (NP_004322), in human. Both proteins contain a BNIP3 domain (PF06553), which covers a BH3 domain and a carboxyl-TM domain. They localize to the mitochondrial outer membrane through their C-terminal TM domains while their N termini are exposed to the cytosol. A short linear motif (SLiM) essential for NIX activity and an LIR motif are labeled with red boxes. The SLiM comprises a triplet of hydrophobic amino acid residues at its center and charged residues on each side of the triplet. Red asterisks indicate conserved sites in LIR, key phosphorylation sites (S17 and S24) in BNIP3, and the central leucine (Leu) residue in the hydrophobic triplet of SLiM in NIX. (D) Alignment of LIR motif and SLiM of BNIP3 and NIX in representative metazoan species. The LIR motif, W(18)-V-E-L(21), in human BNIP3 and its conserved sequences are enclosed within a red box. The SLiM are conserved in vertebrates and are enclosed within a red box. Red asterisks indicate key sites, as labeled in (C). Ola indicates O. latipes and Tru indicates T. rubripes. Information on other species is listed in Table 1. The alignments in (B,D) were colored according to BLOSUM62 score in Jalview (Waterhouse et al., 2009).