Figure 2.
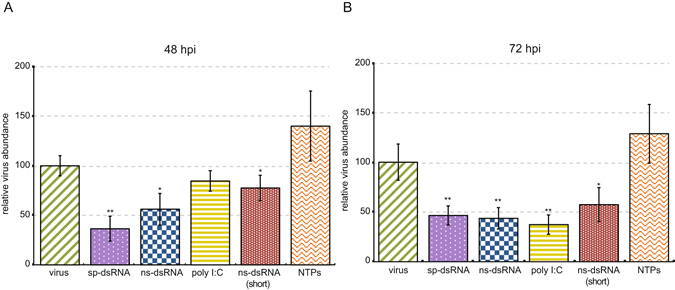
Relative virus RNA abundance was reduced in dsRNA-treated bees as compared to virus-infected bees. Relative abundance of SINV-GFP in individual bees (n = 10) was assessed by qPCR. (A) At 48 hours post-infection (hpi), bees treated with dsRNA (1 kb) had reduced relative virus RNA abundance (includes both virus genomes and transcripts) by 64% for sp-dsRNA-treated bees (dotted purple, **p < 0.005) and 44% for ns-dsRNA-treated bees (checkered blue, *p < 0.05), as compared to bees infected with virus only (green stripes). Likewise, bees treated with short dsRNA (0.5 kb) had 24% less virus (dotted red, *p < 0.05) than virus-infected bees. (B) At 72 hpi bees treated with dsRNA had reduced relative virus abundance by 54% (**p < 0.005) and 56% (**p < 0.005) for sp-dsRNA and ns-dsRNA, respectively. Similarly, bees treated with poly(I:C) (yellow stripes), a structural analog of dsRNA, had 63% less virus than virus-infected bees (**p < 0.005). Bees treated with short dsRNA also had 43% less virus (dotted red, *p < 0.05) than bees infected with virus alone. The virus abundance in bees treated with NTPs (orange wavy lines) was not significantly different from virus-infected bees at either 48 hpi (A) or 72 hpi (B). Percent relative virus abundance for each sample was determined via ΔΔCT analysis (using Am rpl8 as the house keeping gene); statistical differences between treatment and virus-infected bees were performed using one-sided Student’s t-tests, *p ≤ 0.05, **p ≤ 0.005. The bars represent the standard error of the mean.
