Figure 4.
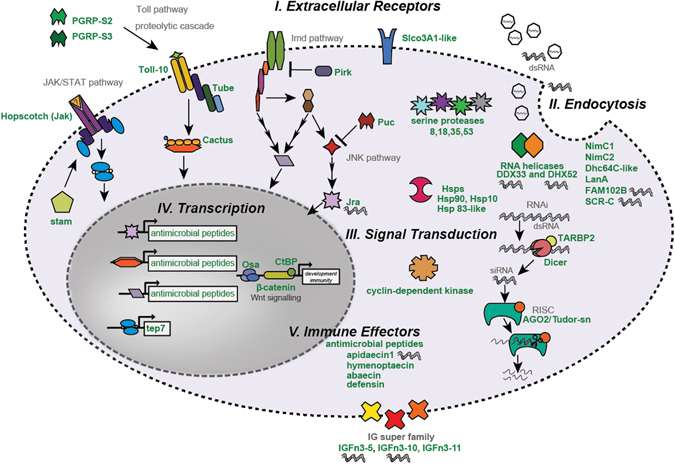
Differentially expressed genes in response to virus and/or dsRNA treatment in a cellular context. The DEGs of virus-infected and dsRNA-treated honey bees were surveyed for differential expression of genes involved in previously characterized insect immune pathways: RNAi, Toll, JAK/STAT (Janus Kinase and Signal Transducer and Activator of Transcription), Imd (Immune Deficiency), and JNK (c-Jun N-terminal kinases)29, 56. This analysis determined that many genes encoding extracellular receptors, proteins involved in endocytosis, signal transduction, and immune responses exhibited increased expression; these DEGs (denoted by bold and green font) are depicted in a cellular context. Many genes exhibited higher fold change in bees treated with both virus and dsRNA as compared to bees infected with virus only (denoted with dsRNA), suggesting their involvement in dsRNA-triggered immune responses. Complete DEG lists are provided in Supplementary Table S6.
