Abstract
Satellite DNAs (satDNAs) constitute large portion of eukaryote genomes, comprising non-protein-coding sequences tandemly repeated. They are mostly found in heterochromatic regions of chromosomes such as around centromere or near telomeres, in intercalary heterochromatin, and often in non-recombining segments of sex chromosomes. We examined the satellitome in the cricket Eneoptera surinamensis (2n = 9, neo-X1X2Y, males) to characterize the molecular evolution of its neo-sex chromosomes. To achieve this, we analyzed illumina reads using graph-based clustering and complementary analyses. We found an unusually high number of 45 families of satDNAs, ranging from 4 bp to 517 bp, accounting for about 14% of the genome and showing different modular structures and high diversity of arrays. FISH mapping revealed that satDNAs are located mostly in C-positive pericentromeric regions of the chromosomes. SatDNAs enrichment was also observed in the neo-sex chromosomes in comparison to autosomes. Especially astonishing accumulation of satDNAs loci was found in the highly differentiated neo-Y, including 39 satDNAs over-represented in this chromosome, which is the greatest satDNAs diversity yet reported for sex chromosomes. Our results suggest possible involvement of satDNAs in genome increasing and in molecular differentiation of the neo-sex chromosomes in this species, contributing to the understanding of sex chromosome composition and evolution in Orthoptera.
Introduction
Among the repetitive sequences in the genomes of eukaryotes, tandem repeats (TRs) are very abundant and are mostly represented by satellite DNAs (satDNAs). SatDNA sequences are mainly located in centromeric, telomeric or intercalary heterochromatin1–3 but, in some cases, also dispersed in euchromatin4, 5. This genomic fraction is composed of hundreds to thousands of noncoding tandemly-arrayed sequences with late-replication, and oriented in a head-to-tail fashion1, 3, 6–9.
SatDNA families, in general, differ in sequence identity, copy number and chromosome distribution2, 10–12. These sequences are subject to intragenomic concerted evolution, resulting in more efficient homogenization of repeats within species than between species and also between repeats located in the same array/chromosome than between different ones2–4, 7. Concerted evolution is achieved through multiple mechanisms of non-reciprocal transfer such as unequal cross-over, gene conversion, rolling-circle replication and transposition13, 14.
Sex chromosomes have arisen independently several times in a wide range of animals and plants from an ordinary autosomal pair15, 16, presenting as a recurrent trait the suppression of recombination and accumulation of distinct classes of repetitive DNAs, including satDNAs1, 17–21. In Orthoptera, the X0♂/XX♀ sex-determining system is considered modal22, 23 but eventually, diverse sex chromosome systems evolved several times, such as neo-XY♂/XX♀24–26, X1X20♂/X1X1X2X2♀27 and even neo-X1X2Y♂/X1X1X2X2♀25, 28. It was found that particularly centric fusions (i.e. Robertsonian translocations) and tandem fusions with autosomes, dissociations and inversions contributed to the formation of neo-sex chromosomes in Orthoptera 22, 23, 25–28. The DNA composition of the orhtopteran neo-sex chromosomes was studied only in a few species, mainly focusing on certain types of repetitive DNAs, such as multigene families, C0 t DNA fraction, telomeric repeats and microsatellites arrays25–29.
Chromosomal evolution and repetitive DNA organization was addressed in the cricket Eneoptera surinamensis, a species with the genome size of 5.42 Gbp, chromosome number of 2n = 9♂/10♀ and a neo-X1X2Y♂/X1X1X2X2♀ sex-determining system28. During male meiosis the multiple sex chromosomes remain unpaired and do not form chiasmata, suggesting that they do not proceed recombination22, 23. The neo-Y chromosome is the largest element and it exhibits multiple heterochromatic bands, while the neo-X1 and neo-X2 are poor in C-positive heterochromatin. Of the repetitive DNAs, two multigene families (5S rDNA and histone genes), C0 t DNA fraction, and diverse microsatellites mapped to the neo-Y28.
In the present study, in order to provide comprehensive information on repetitive DNAs that occur in the genome, and specifically in the neo-sex chromosomes of E. surinamensis, we performed a detailed analysis of satDNAs (the satellitome)30, integrating genomic and chromosomal data. Our results revealed accumulation of satDNAs, with the occurrence of unusually high number of 45 distinct families in the E. surinamensis genome. In addition, some of these families were enriched in the highly differentiated neo-X1X2Y sex chromosomes in comparison to autosomes, mostly over-represented in the neo-Y. To our knowledge, this is the largest diversity of satDNAs yet reported for a Y chromosome in eukaryotes.
Results
Identification of satDNAs and sequence characterization
Illumina sequencing returned 5,872,912 paired-end reads (ranging from 35 to 288 nt, mean reads length of 164.6 nt) totaling 615,581,150 nt that were trimmed to 150 nt. Given the estimated genome size of 5.42 Gbp for E. surinamensis 28, this represents about 0.16× genome coverage. The clustering analysis through the RepeatExplorer31, 32 used as input 1,299,110 illumina previously trimmed paired-ends reads and produced 292,070 clusters (containing 92.3% of reads) differing in size, sequence composition and genomic abundance, including satDNAs and other non-characterized repetitive elements. The singletons should represent the low copy number fraction of the genome, which yielded in 100,002 singlets (containing 7.7% of reads) (Fig. 1a). A set of 131 of the most abundant clusters representing repetitive elements was analyzed in the search for satDNAs. A representation of the genomic proportion of 45 clusters identified as satDNAs is showed in Fig. 1b.
Figure 1.
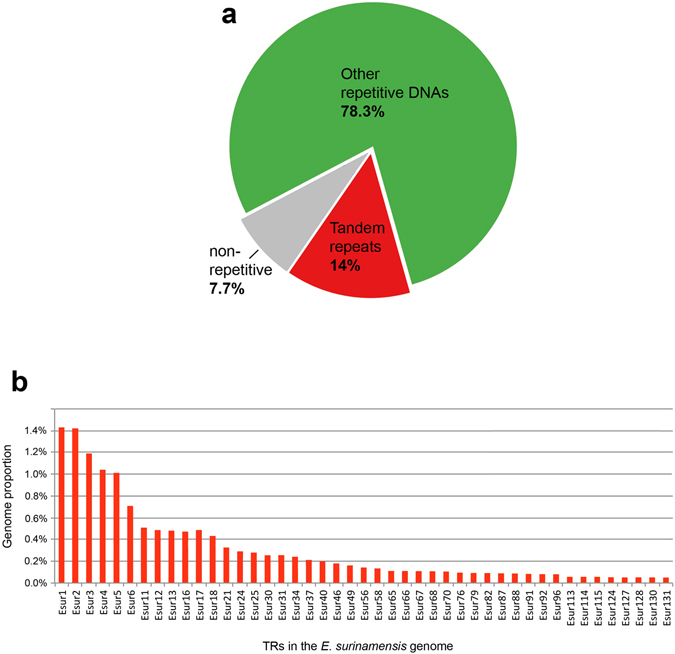
(a) Overview of the repetitive composition of E. surinamensis genome based on the output of RepeatExplorer. (b) Genomic proportion for each of the 45 satDNAs isolated and characterized. TRs, tandem repeats.
The analysis of dotplots confirmed the occurrence of 45 families of tandem repeats ranging from 4 to 517 bp long, showing different modular structure and array diversity. Among them, 21 satDNAs families were recovered through PCR that revealed a ladder pattern. For the remaining repeat families, with small monomers (less than 60 bp), we were not able to confirm the ladder pattern through PCR due to the very limited window for primer design and amplification. The characteristics for all satDNAs families are presented in the Supplementary Tables S1 and S2. Altogether, the 45 satDNAs families comprised about 14% of the male genome that represent 758,8 Mb of 5,420,00 Mb of species genome size. The nucleotide divergence within the families varied from 1.9 to 30.2%. The nucleotide sequences showed an A + T content ranging from 15 to 67.15%. It can be concluded that most of the satDNAs analyzed here constitute light satDNAs due to the low A + T content (see Supplementary Tables S1 and S2).
For two satDNAs, i.e. Esur17 and Esur18, two subfamilies were identified showing an individual repeat called α and a composite repeat called α/β (see Supplementary Table S2 and Supplementary Fig. S1). There was no significant sequence similarity between α and β sequences. The size of the Esur17 α-repeat was 163 bp while the Esur17 α/β composite repeat was 317 bp, each consisting of an α-unit and additional 154 bp β-unit. The nucleotide divergence between both Esur17-α and Esur17-α/β repeats was 9.4% (see Supplementary Table S2). Regarding Esur18, our sequences analysis showed that this is an array composed of α/β composite units with 517 bp, consisting of a 329 bp α-unit and a 188 bp β-unit (see Supplementary Fig. S1). The Esur18 α/β repeats showed 4.3% nucleotide divergence (see Supplementary Table S2). We also detected sequence similarity between Esur2 and Esur34 repeats (~82.6%) and between Esur3 and Esur58 repeats (80.9%). This result explains the overlapping chromosomal distribution of both satDNA pairs (see below) and indicate that they are representatives of two satDNA families, Esur2/Esur34 and Esur3/Esur58. NJ trees showed Esur2, Esur34, Esur3 and Esur58 allocated in cluster-specific branches, indicating that each subfamily is composed of exclusive repeat-variants (see Supplementary Fig. S2). Both NCBI BLAST and Repbase searches, with the consensus monomer sequence belonging to each repeat family as a query, did not revealed significant similarity with any other previously described sequences.
Considering all satDNAs identified, we found that monomer size and nucleotide divergence displayed a somewhat low but significant negative correlation (rho = −0.48, P = <0.001). Also, monomer size and a total number of loci displayed a moderate negative correlation (rho = −0.63, P = <0.0001) (Supplementary Fig. S3).
Chromosomal localization of satDNAs
To detect the chromosomal localization of the 45 satDNA families single- or two-color FISH were carried out on male mitotic metaphases. In autosomes, distinct satDNAs were located mostly in the pericentromeric regions that correspond to the C-band positive blocks observed by Palacios-Gimenez et al.28. A few satDNA loci were also found in interstitial and distal C-band negative blocks. The patterns were variable depending on the repeat mapped (Fig. 2, Supplementary Tables S1 and S2). Most satDNAs were located in multiples autosomes, while some of them were located exclusively in one chromosome pair, for example, Esur4, Esur6, and Esur18- α/β, were all co-located in the centromere and secondary constriction of pair 1. Two-color fiber-FISH confirmed regions of interspersion between Esur4, Esur6 and Esur18- α/β repeats (see Supplementary Fig. S4). For Esur17, the α and α/β repeats were located in the secondary constriction of pair 1 and in the X2 chromosome; for Esur18, the α and β repeats were located in secondary constriction of pair 1, but additional multiple loci of the α repeats were detected in the Y chromosome (see Supplementary Table S2 and Supplementary Fig. S5).
Figure 2.
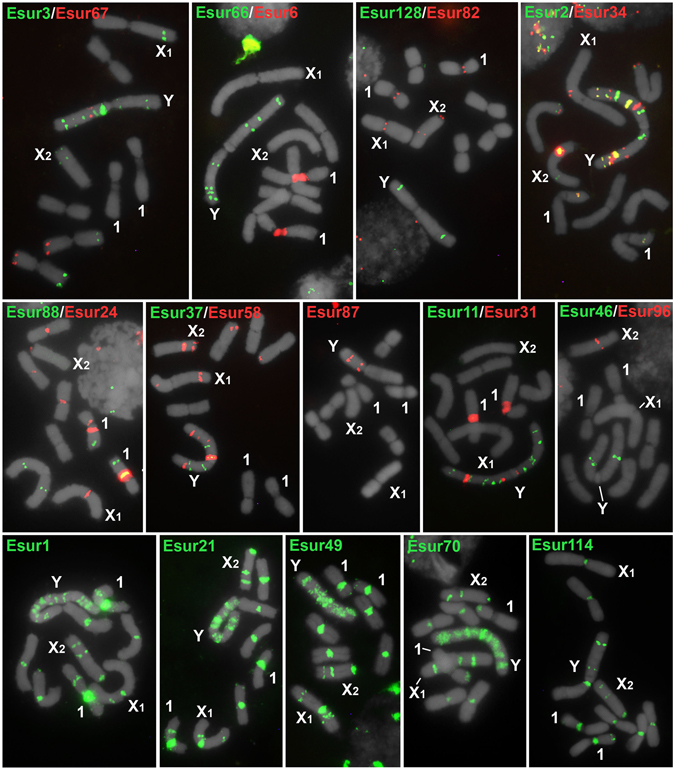
Chromosomal location of 22 satDNAs in mitotic chromosomes of male embryos of E. surinamensis by FISH. Upper and middle panels show satDNAs with monomers >60 bp, lower panels satDNAs with monomers <60 bp. The satDNA family names and hybridization signals for each type of the probe are shown in the images by colors. Note the enrichment in the neo-Y (Y) sex chromosome with multiple loci or spread signals for some repeats, while the X1 and X2 sex chromosomes show only a few or even none hybridization signals.
Some of satDNAs families were placed in the neo-X1 and neo-X2 but the abundance (number of loci) of satDNAs on the neo-Y chromosomes was remarkable, showing either multiple discrete loci or a scattered pattern (Fig. 3). The abundance in relation to the number of loci on the neo-Y chromosome was also noteworthy compared to autosomes and the neo-X1 and neo-X2 chromosomes (Fig. 2 and Supplementary Tables S1 and S2). Among the 45 satDNAs families, 39 were located in the neo-Y chromosome and six of them (Esur11, Esur31, Esur37, Esur46, Esur65 and Esur66) through FISH, mapped exclusively to the neo-Y chromosome, however, they were recovered through PCR in both sexes (Fig. 3, Supplementary Tables S1 and S2).
Figure 3.
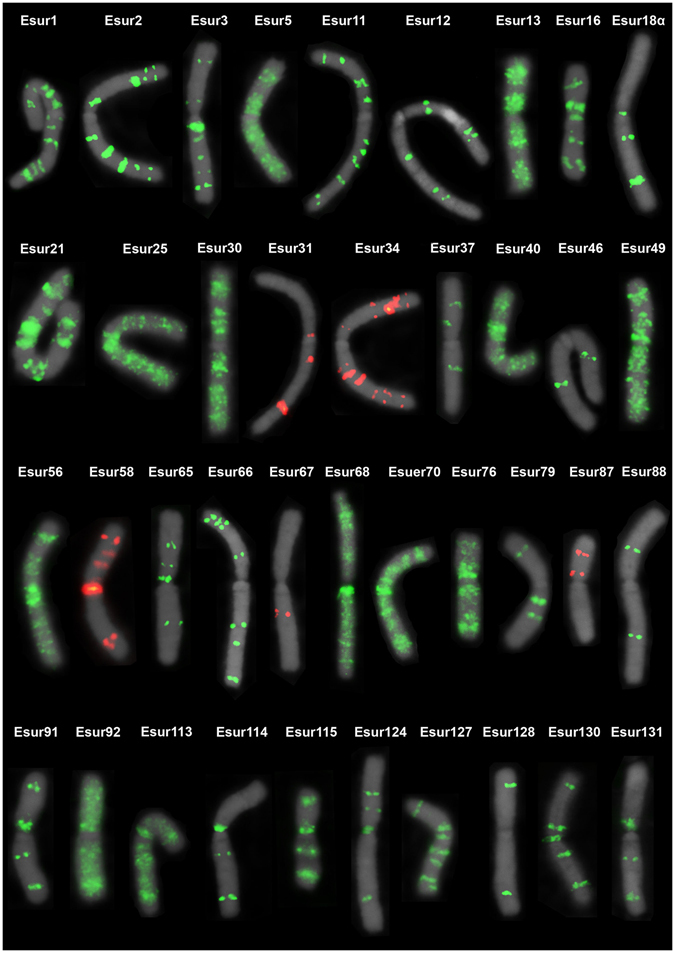
Distribution of the 39 satDNAs in the neo-Y chromosome of E. surinamensis.
Male versus female satDNAs abundance
The comparative relative abundance of satDNAs between male and female genomes carried out through qPCR clearly show that the satDNAs doses differ significantly between sexes, with males harboring a higher copy number than females for most of repeats studied (Fig. 4, Supplementary Table S3). For example, most remarkable difference were seen in Esur2, Esur11, Esur46, Esur65, Esur66, Esur87 and Esur128, with males showing six to ten times more copies than females (Fig. 4).
Figure 4.
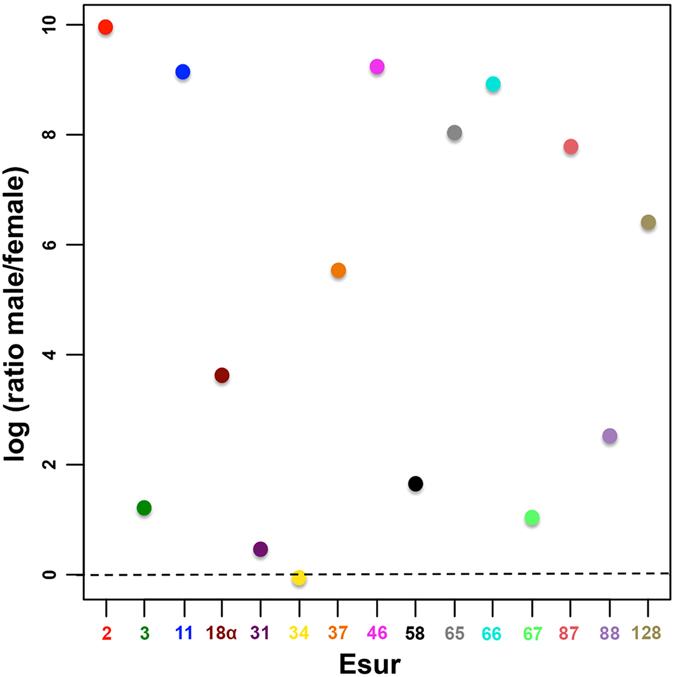
Copy number variation between male vs female genomes of 15 satDNAs with monomers >60 bp in E. surinamensis. The male/female ratio of a relative copy number is shown on a logarithmic scale. The qPCR of male and female genomic DNA was used to calculate the satDNA dose by a ΔCt method of relative quantification (see Supplementary Table S3). Each satDNA family (Esur) is represented by numbers and colors directly in the images. Note that most satDNAs differ significantly between sexes with males having a higher copy number than females for most repeats studied. The statistical significance for difference in copy number between males and females using chi-squared test was highly significant, p-value < 0.01 for each satDNA.
Discussion
General organization of satDNAs in the genome of E. surinamensis
Through the graph-based clustering of sequencing reads of the E. surinamensis genome, followed by complementary analysis, we discovered 45 new satDNAs families. These satDNAs coexist in the E. surinamensis genome, with variable monomer sizes and nucleotide divergence, accounting for ~14% of the genome (758.8 Mb). It is well known that diverse satDNA families are commonly found in eukaryotic genomes, e.g., in Olea europea with six satDNA families33, Tribolium castaneum with nine34, and Camellia japonica with four35, reaching up to 16 satDNAs in Drosophila melanogaster (reviewed by ref. 36) and 62 in the grasshopper L. migratoria 30. Besides the considerable proportion of satDNAs in the genome of E. surinamensis, it is also remarkable the presence of so many different families in comparison with most eukaryotes. Another interesting feature of E. surinamensis is that no satDNA family is predominant, which contrasts with what has been observed in several other species, in which a few families prevailed33–36.
We could speculate that the accumulation of satDNAs in E. surinamensis could have been favored by highly rearranged karyotype (2n = 9♂) in comparison with the modal diploid chromosome number in Gryllidae crickets23. Alternatively, the high diversity of repetitive DNA itself might facilitate/cause such a complex karyotype rearrangements. It is well known that chromosomal rearrangements may involve highly repetitive DNA sequences, since these could provide sites for karyotype reshuffling without detrimental effects on the integrity of coding sequences37–39. In any case, the rearranged karyotype of E. surinamensis represents a specific environment with limited recombination that could facilitate the rate of homogenization of repetitive DNA, like in multiple sex chromosomes (see below). In addition to chromosome rearrangements, mechanisms involved in the satDNAs evolution could have occurred, such as amplification mediated by rolling-circle replication and reinsertion, unequal crossing-over between DNA repeats from sister chromatids, transposition and gene conversion. These mechanisms have been suggested as possible causes of sequence homogenization within satDNA1, 8. The putative involvement of highly rearranged karyotype for satDNA multiplication could be supported by comparative analysis with the related species Gryllus assimilis (Gryllidae) with higher diploid number that is ancient for cricket, in which the satDNA analysis revealed the occurrence of only 11 satDNA families (Palacios-Gimenez et al., submitted). Considering the genome size (G. assimilis 2.13 Gb and E. surinamensis 5.42 Gb) and quantity of satDNA in each species (G. assimilis 4% and E. surinamensis 14%) the quantity of satDNAs was increased about 8.9 times in the genome of E. surinamensis.
We found that most satDNAs are non-homologous, suggesting inefficient homogenization between distinct repeats. The intra-family homogenization is evident, even more for larger satDNAs than smaller ones, and could be directly related to the sequence size and specific chromosomal distribution. Comparing the distribution of the large- and small-sized satDNAs it is evident that the smaller, ranching up to 60 bp, repeats are more dispersed than larger ones (Fig. 2), mostly occurring in all chromosomes of the complement, which could facilitate their sequence divergence.
In our data, the satDNA monomer size was negatively correlated with sequence divergence (rho = −0.48, P < 0.001) (Supplementary Fig. S3). A similar trend was also observed for three satDNAs of the cave cricket Dolichopoda schiavazzii 40. This finding disagrees with predictions from computer simulations, which anticipated a direct correlation between the monomer size and the divergence due to low recombination frequency41. However, as noted by Bachmann et al.40, A + T content and sequence complexity, which were not included in these early models, could play a role in facilitating recombination and homogenization of the repeats. We also found a negative correlation between the satDNA monomer size and the number of loci (rho = −0.63, P < 0.0001) (Supplementary Fig. S3). The distribution of small satDNAs in almost all chromosomes, and remarkably in the neo-Y, could be the result of dispersion through extra-chromosomal circular DNAs, formed via intrastrand recombination, and subsequent reintegration elsewhere in the genome by illegitimate or homologous recombination (reviewed in ref. 42). In addition, the movement of otherwise non-transposable DNA fragments via their capture by transposable elements could represent another possible mechanism of the intragenomic dispersion of tandem repeats43–47. What remains is to investigate the exact role played by the monomer size in facilitating satDNA dispersion.
For the large satDNAs, which exhibit in a more chromosome-specific distribution, our combined analysis using cytogenetic and genomic tools revealed a distinct degree of complexity, i.e., simple tandem arrangements and composed units, like α/β repeats for Esur17 and Esur18. satDNAs with composite units have been reported for example in Chrysolina carnifex 48 Tribolium brevicornis 49 and primates50. Besides the composite units observed for Esur17 and Esur18 we also found cases of overlapping distribution of satDNAs as demonstrated by fiber-FISH for the satDNAs families exclusively located in chromosome pair 1, Esur4, Esur6 and Esur18. It occurred independently of sequence homology. Moreover, we observed other cases of co-localization depending on the chromosome. Overlapping distribution of satDNAs is usually reported for repeats with a remarkable similarity, such as the satellite I and satellite II subfamilies in Tribolium madens 51 the pBuM-1 and pBuM-2 subfamilies in the Drosophila buzzatii species complex52 and the psr2 and psr18 subfamilies in Nasonia vitripennis 53. However, overlapping distribution of non-homologous satDNAs as observed here for E. surinamensis was reported only in a few cases, e.g., the DBC-150 and pBuM satDNAs family in several Drosophila species54 the pSc200 and pSC250 families in the rye chromosomes55 and the PROsat, PSUchr1sat and PsatDNA satDNAs in two species of hamsters56.
The complexity of the evolution of satDNAs identified in the E. surinamensis genome is reflected by (i) junctions of monomers forming complex units, as explained above and reinforced (ii) by the occurrence of divergent satDNAs, probably originating from a common ancestor, as indicated by the sequence similarity. For example, we found an apparent similarity of monomer satDNA sequences between Esur2 and Esur34 and between Esur3 and Esur58. However, each of these sequences in the constructed NJ tree constitutes separate branches. This allows us to conclude that these sequences belong to closely related satDNAs families with a common ancestor but they diverged during their evolution, including moving to distinct chromosomal regions. Moreover, these sequences differentially amplified which resulted in variation in their copy number, as demonstrated by qPCR experiments.
Accumulation of satDNAs occurred in the E. surinamensis neo-Y chromosome
It is well known that the Y or W sex chromosomes accumulate high quantities of distinct classes of repetitive DNAs due to the low frequency of recombination1, 18, 19, 21, 57, 58. In some species, it was shown that these repetitive sequences are involved in genetic degeneration, chromatin organization and regulation of expression36, 57, 59. This could also be the case in the E. surinamensis sex chromosomes that remain either unpaired and achiasmatic during male meiosis, suggesting the absence of recombination between them22, 23. Regarding satDNAs, more than one family harboring sex chromosomes were reported, for example, in D. melanogaster 60, Muntiacus muntjac 61, 62, T. castaneum 34, Rumex acetosa 63–65 and Silene latifolia 66. However, in comparison with these species the E. surinamensis neo-Y chromosome harbors the highest diversity of satDNAs documented to date, representing 39 distinct families, with seven being exclusive to this chromosome. Moreover, most satDNAs are enriched in this sex chromosome and constitute multiple loci as shown by FISH. These findings suggest that the neo-Y accumulated satDNAs after its origin, causing its enlargement, in comparison with the neo-X1 and neo-X2. A similar pattern was reported for the large Y chromosomes of plants that also accumulated repetitive DNAs65, 66 in contrast to the small mammalian Y chromosomes67, 68.
Our data suggest a complex origin of the neo-sex chromosomes in E. surinamensis, involving more than two simple translocations anticipated for the Orthoptera neo-X1X2Y sex chromosomes22, 23, 25. The evolution of E. surinamensis sex chromosomes probably included centric and tandem fusions and inversions, like in M. muntjac 61, 62. The multiple interstitial satDNAs loci in the E. surinamensis neo-Y could represent remnants of the ancestral centromeric material at chromosome fusions sites, or they are the consequence of massive expansion of DNA repeats that could induce new chromosomal rearrangements.
Although some satDNA families were exclusively mapped by FISH to the neo-Y chromosome, they were also recovered in the female genomic DNA using PCR, indicating their occurrence in other chromosomes, at least in a low copy number. Differential expansion of satDNAs between sexes was corroborated by qPCR. Our results clearly showed that the doses of most satDNAs examined differ significantly between sexes, probably due to differential expansion, with males having on average two to ten times more copies than females. We hypothesize that the differential repeats distribution patterns on the E. surinamensis sex chromosomes can be explained by a higher rate of colonization and insertion and lower rate of removal of satDNAs in the neo-Y chromosome, similarly to that suggested for the R. acestosa sex chromosomes65. Therefore, these satDNAs lead to compartmentalization of the neo-Y chromosome and a chromosome-wide DNA sequence diversity. In addition, the neo-Y chromosome may exhibit a differentiated long-range chromatin structure/composition compared to autosomes and both neo-X chromosomes. Partially-heterochromatic appearance of the neo-Y chromosome in E. surinamensis with G + C-rich blocks dispersed in its entire length25 supports this view. The involvement of neo-sex chromosomes in increasing of satDNAs quantity and diversity in E. surinamensis emerges by analysis of G. assimilis with X0 sex system, which is ancestral for Orthoptera as a whole22, 23. In this species only 11 satDNAs families were recovered by the same papiline used here. Among them only eight populate the sex chromosome with discrete bands mainly in terminal region (Palacios-Gimenez et al. submitted).
Our data, together with the analysis of sex chromosome that revealed either unpaired and achiasmatic during meiosis22, 23 − a common feature for old sex chromosomes69 − suggest the occurrence of old neo-sex chromosome system in E. surinamensis, which contrasts with the described young sex chromosomes21, 70, 71. Moreover considerable accumulation of various satDNAs and other repetitive DNA classes in the neo-Y chromosome25 reinforce this view, as reported in other species67, 69. This could mean that there are no evolutionary strata on the E. surinamensis sex chromosomes, similar to the strata found in human X chromosome72. Theoretically, satDNAs should accumulate more intensively in a region of the Y chromosome corresponding to the ancestral X chromosome, in which recombination was ceased earlier, but no such region was identified in our study, supporting this view.
Our study provides important information concerning composition and evolution of neo-sex chromosomes among crickets, suggesting the involvement of neo-sex chromosomes in amplification and sequence divergence for satDNAs, which generated the highest diversity of satDNAs in sex chromosomes. This data with future analysis using other repetitive sequences and sex related single copy genes will be also relevant to understand precise composition of sex chromosomes, helping in the understanding of sex chromosome evolution, sex determination and possible mechanisms involved in dosage compensation, an issue almost completely unknown in Orthoptera. Moreover to shed light in the evolutionary history of sex chromosomes in crickets it will be informative the analysis of other species with divergent sex systems using similar strategies applied here in a comparative manner.
Materials and Methods
Samples, Chromosome Preparations and Genomic DNA Extraction
Males and females of Eneoptera surinamensis were collected in the Parque Estadual Edmundo Navarro de Andrade (Rio Claro, SP, Brazil) between May 2013 and March 2014 with the authorization of COTEC (process number 341/2013) and were maintained in captivity until oviposition. Mitotic chromosomes preparations were obtained from embryo neuroblasts using standard procedures described elsewhere73. In addition, adult male testes were dissected and fixed in Carnoy’s modified solution (3:1, 100% ethanol: absolute acetic acid). Genomic DNA of adult males and females were extracted from femurs using the phenol/chloroform-based procedure described in Sambrook and Russel74.
Illumina sequencing and graph-based clustering of sequencing reads
Paired-ends sequencing (2 × 300) was applied in libraries prepared as recommended by illumina (illumina Inc., San Diego, CA, USA) using Nextera DNA Library Preparation Kit v3 to one male specimen genomic DNA. Library fragments sizes were in the range from 400 to 600 bp and sequencing was performed using Miseq Sequencing System. The obtained reads were preprocessed to check the quality of the reads with FASTQC75 and we did a quality filtering with the FASTX-Toolkit suit76. The paired-end reads were also trimmed at 150 nt in length, and then were joined using the “fastq-join” software of the FASTX-Toolkit suit76 using default options. Based on illumina sequencing we estimated the G + C content of the whole genome using FastQC High Throughput Sequences QC Report version: 0.11.4 (available at http://www.bioinformatics.babraham.ac.uk/projects/). To search for satDNAs in the E. surinamensis genome, we carried out a graph-based clustering and assembly of these sequences using the RepeatExplorer31, 32. Afterward, we searched for clusters that showed repeat graph density in summary output, which is a typical characteristic of satDNAs families in this approach31, and refined this search using Dot plot charts implemented in Dotlet77.
Isolation and sequence analysis of satDNAs
Clusters with high graph density were submitted to the Tandem Repeats Finder (TRF) algorithm78 to identify the DNA sequence that maximized the alignment scores between the different monomers that could be defined in tandem. The TRF alignment parameters were 2, 3, 5 for match, mismatch and indels, respectively, and a minimum alignment score of 50 was required for reporting. Additionally, we used the dotplot graphic alignment tool implemented in Dotlet77 to identify monomers of the same family and to confirm the tandem organization. The monomers with maximum length were used as the representative copy for each satDNA family, and as the query sequences for further BLAST (http://www.ncbi.nlm.gov/Blast/) and Repbase (http://www.girinst.org/repbase/) searches to check similarity with published sequences. Sequence alignments of satDNAs copies were performed using Muscle79 implemented in MEGA580. MEGA5 was also used to estimate nucleotide divergence (p distance), A + T content and perform repeat length analysis. The evolutionary relationships among sequences were inferred by neighbor-joining (NJ) trees using the implemented option in MEGA5 and the proportion of nucleotide differences (p distance). Satellite alignments are available upon request to the author.
The consensus sequences of each satDNA family was used to design primers with opposite directions (Supplementary Table S4), using the Primer3 software81 or manually. In order to verify the presence of satDNAs families in male and female, we performed polymerase chain reactions (PCR). PCRs were carried out using 10× PCR Rxn Buffer, 0.2 mM MgCl2, 0.16 mM dNTPs, 2 mM of each primer, 1 U of Taq Platinum DNA Polymerase (Invitrogen, San Diego, CA, USA) and 50–100 ng/μl of template DNA. The PCR conditions included an initial denaturation at 94 °C for 5 min and 30 cycles at 94 °C (30 s), 55 °C (30 s), and 72 °C (80 s), plus a final extension at 72 °C for 5 min. The PCR products were visualized on a 1% agarose gel. The monomeric bands were isolated and purified using the Zymoclean™ Gel DNA Recovery Kit (Zymo Research Corp., The Epigenetics Company, USA) according to the manufacturer’s recommendations and then used as source for reamplification.
To check the isolated sequences, the purified PCR products were sequenced in both directions using the service of the Macrogen Inc., and then compared to the consensus sequences obtained by the genomic analysis. The consensus sequences for each satDNAs family can be found in Supplementary Results S1, and sequence alignments are available upon request.
Probes and fluorescence in situ hybridization (FISH)
PCR products for each satDNA family were labeled by nick translation using biotin-14-dATP (Invitrogen) or digoxigenin-11-dUTP (Roche, Mannheim, Germany). SatDNAs with less than 60 bp were labelled directly at the 5′ end with biotin-14 dATP (Sigma-Aldrich, St Louis, MO, USA) during their synthesis. Single or two-color FISH was carried out according to Pinkel et al.82 with modifications83 using mitotic chromosome preparations. Fiber-FISH experiments were conducted as described in de Barros et al.84 using suspensions of testis cells. The probes that were labeled with digoxigenin-11-dUTP were detected using anti-digoxigenin-rhodamine (Roche) and the probes labeled with biotin-14-dATP were detected using streptavidin conjugated with Alexa Fluor 488 (Invitrogen).
Following FISH, chromosomal preparations were counterstained using 4′,6-diamidine-2′-phenylindole (DAPI) and mounted in VECTASHIELD (Vector, Burlingame, CA, USA). Chromosomes and hybridization signals were observed using an Olympus BX61 fluorescence microscope equipped with appropriate filter sets. Black-and-white images were recorded using a DP71 cooled digital camera. The images were pseudo-colored in blue (chromosomes) and red or green (signals), merged and optimized for brightness and contrast using Adobe Photoshop CS2.
Quantitative analysis of satDNAs
Quantitative PCR (qPCR) using male and female genomic DNA as templates was used to check the copy number differences between males and females of selected satDNAs, i.e., some of the repetitive families larger than 60 bp length (see primers in Supplementary Table S4). The selected satDNAs were chosen due to their presence on the neo-Y chromosome determined by FISH. The qPCR of male and female genomic DNA was used to calculate the satDNA dose by a ΔCt method of relative quantification85. Gene dosage ratios (GDR) of the target satDNAs were compared with a reference gene, 70-kDa heat shock protein (Hsp-70) family using as primers F: 5′-GGTGGTATGACCACTCTTATCAA-3′ and R: 5′-CACTTCATTTTGAGGCACACC-3′ that were designed according to the Hsp-70 gene from Locusta migratoria (accession number AY178988). This gene was used as a reference because there were no differences detected in amplification rates between sexes in E. surinamensis, suggesting that the Hsp-70 gene is autosomal and has equal copy number in both sexes. Because we were not sure whether Hsp-70 is a single-copy gene, the quantitative analysis was a relative comparison of the gene dose between male and female. Quantitative analyses were carried out in MicroAmp® Fast Optical 96-Well Reaction Plate with Barcode (0.1 mL) (Applied Biosystems, Life Technology™, Carlsbad, CA) covered by Optical Adhesive Covers (Applied Biosystems) using the StepOne Real-Time PCR Systems Thermal cycler. qPCR in both the target satDNAs and the reference gene were performed simultaneously in triplicates of three independent samples, i.e. 3 males and 3 females. Each qPCR mixture contained 6.25 μl 2× GoTaq® qPCR master mix (Promega, Madison, WI, USA), 0.25 mM of each forward and reverse primers and 30 ng of either male or female genomic DNA, in a final volume of 10 μl. qPCR mixtures without DNA served as negative controls. The cycling conditions were 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Specificity of the PCR products was confirmed by analysis of the dissociation curve. A correlation analysis of GDR means of target satDNAs between males and females were carried out with R statistical software version 3.3.186 and edited using Adobe Photoshop CS2.
Statistic correlation analysis
We used the PerformanceAnalytics package87 implemented in the R statistical software version 3.3.186 to calculate Spearman’s rank correlation coefficients for satDNA monomer size, genome proportion, A + T content, nucleotide divergence and loci number. For the satDNAs showing spread FISH signals (see Supplementary Tables S1 and S2), the total countable loci number was increased by 10 to account for their dispersed profile, but still in a conservative manner to avoid counting the possible existing nonspecific hybridization signals.
Electronic supplementary material
Acknowledgements
The authors are grateful to “Parque Estadual Edmundo Navarro de Andrade” administration for sample collecting authorization and Antonio Sergio Pascon for technical assistance in obtaining embryos. We thank Dr. Frantisek Marec (Institute of Entomology, Biology Centre CAS, České Budějovice, Czech Republic) for the critical review of a previous version of this manuscript, and Dr. Melvin Bonilla (Harvard T.H Chan School of Public Health, Boston, MA, USA) for English corrections. To the anonymous reviewers for valuable suggestions. OMPG acknowledges scholarship obtained from Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP (process number 2014/02038-8). This study was supported by FAPESP (process number 2014/11763-8) and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior-CAPES. DCCM was awarded a research productivity fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (process number 304758/2014-0).
Author Contributions
O.M.P.G. designed the experiments, collected specimens and harvest tissues, performed the FISH mapping and the bioinformatics analyses, interpreted the data and drafted the text. G.B.D., L.G.L. and G.C.S.K. performed the bioinformatics analyses and interpreted the data. E.R. and C.M. prepared nucleic acids, sequencing libraries and interpreted the data. D.C.C.M. conceived and designed the experiments, analyzed the data and drafted the text. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-06822-8
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 2.Plohl M, Luchetti A, Meštrović N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 3.López-Flores, I. & Garrido-Ramos, M. A. Repetitive DNA in The Repetitive DNA Content of Eukaryotic Genomes (ed. Garrido-Ramos, M. A.) 1–28 (Genome Dyn. Basel, Karger, vol 7 (2012). [DOI] [PubMed]
- 4.Kuhn GCS, Küttler H, Moreira-Filho O, Heslop-Harrison JS. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Mol. Biol. Evol. 2012;29:7–11. doi: 10.1093/molbev/msr173. [DOI] [PubMed] [Google Scholar]
- 5.Larracuente AM. The organization and evolution of the Responder satellite in species of the Drosophila melanogaster group: dynamic evolution of a target of meiotic drive. BMC Evol. Biol. 2014;14:233. doi: 10.1186/s12862-014-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large-scale organization of plant chromosomes. Trends in Plant Sci. 1998;3:195–199. doi: 10.1016/S1360-1385(98)01223-0. [DOI] [Google Scholar]
- 7.Palomeque T, Lorite P. Satellite DNA in insects: a review. Heredity. 2008;100:564–573. doi: 10.1038/hdy.2008.24. [DOI] [PubMed] [Google Scholar]
- 8.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Lorenzen MD, Beeman RW, Brown SJ. Analysis of repetitive DNA distribution patterns in the Tribolium castaneum genome. Genome Biol. 2008;9:R61. doi: 10.1186/gb-2008-9-3-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King K, Jobst J, Hemleben V. Differential homogenisation and amplification of two satellite DNAs in the genus Cucurbita (Cucurbitaceae) J. Mol. Evol. 1995;41:996–1005. doi: 10.1007/BF00173181. [DOI] [PubMed] [Google Scholar]
- 11.Wang ZX, Kurata N, Saji S, Katayose Y, Minobe Y. A chromosome 5-specific repetitive DNA-sequence in rice (Oryza sativa L.) Theor. Appl. Genet. 1995;90:907–913. doi: 10.1007/BF00222902. [DOI] [PubMed] [Google Scholar]
- 12.Vershinin AV, Heslop-Harrison JS. Comparative analysis of the nucleosomal structure of rye, wheat and their relatives. Plant Mol. Biol. 1998;36:149–161. doi: 10.1023/A:1005912822671. [DOI] [PubMed] [Google Scholar]
- 13.Dover GA. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- 14.Dover G. Molecular drive. Trends Genet. 2002;18:587–589. doi: 10.1016/S0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- 15.Ohno, S. Sex Chromosomes and sex linked genes, Springer, Berlin (1967).
- 16.Bull, J. J. Evolution of Sex Determining Mechanisms, Menlo Park, CA, Benjamin Cummings (1983).
- 17.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. doi: 10.2307/1312947. [DOI] [Google Scholar]
- 18.Steinemann M, Steinemann S. The enigma of Y chromosome degeneration: TRAM, a novel retrotransposon is preferentially located on the neo-Y chromosome of Drosophila miranda. Genetics. 1997;145:261–266. doi: 10.1093/genetics/145.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinemann S, Steinemann M. Retroelements: tools for sex chromosome evolution. Cytogenet. Genome Res. 2005;110:134–143. doi: 10.1159/000084945. [DOI] [PubMed] [Google Scholar]
- 20.Bachtrog D. A dynamic view of sex chromosome evolution. Curr. Opin. Genetic Dev. 2006;16:578–85. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser VB, Bachtrog D. 2010. Evolutions of sex chromosome in insects. Annu. Rev. Genet. 2010;44:91–112. doi: 10.1146/annurev-genet-102209-163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White, M. J. D. Animal cytology and evolution, Cambridge: Cambridge University Press (1997).
- 23.Hewitt, G. M. Grasshoppers and crickets, Animal Cytogenetics, vol 3, Insecta 1, Orthoptera, Gebrüder Borntraeger, Berlin (1979).
- 24.Mesa A, Fontanetti CS, García Novo P. Does an X-autosome centric fusion in Acrdoidea condemn the species to extinction? J. Orthoptera Res. 2001;10:141–146. doi: 10.1665/1082-6467(2001)010[0141:DAXACF]2.0.CO;2. [DOI] [Google Scholar]
- 25.Palacios-Gimenez OM, Castillo ER, Martí DA, Cabral-de-Mello DC. Tracking the evolution of sex chromosome systems in Melanoplinae grasshoppers through chromosomal mapping of repetitive DNA sequences. BMC Evol. Biol. 2013;13:167. doi: 10.1186/1471-2148-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios-Gimenez OM, Marti DA, Cabral-de-Mello DC. Neo-sex chromosomes of Ronderosia bergi: insight into the evolution of sex chromosomes in grasshoppers. Chromosoma. 2015;124:353–365. doi: 10.1007/s00412-015-0505-1. [DOI] [PubMed] [Google Scholar]
- 27.Palacios-Gimenez OM, Cabral-de-Mello DC. Repetitive DNA chromosomal organization in the cricket Cycloptiloides americanus: a case of the unusual X1X20 sex chromosome system in Orthoptera. Mol. Genet. Genomics. 2015;290:623–631. doi: 10.1007/s00438-014-0947-9. [DOI] [PubMed] [Google Scholar]
- 28.Palacios-Gimenez OM, Carvalho CR, Ferrari Soares FA, Cabral-de-Mello DC. Contrasting the chromosomal organization of repetitive DNAs in two Gryllidae crickets with highly divergent karyotypes. PLoS ONE. 2015;10:e0143540. doi: 10.1371/journal.pone.0143540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugrov AG, Jetybayev IE, Karagyan GH, Rubtsov NB. Sex chromosome diversity in Armenian toad grasshoppers (Orthoptera, Acridoidea, Pamphagidae) Comp. Cytogen. 2016;10:45–59. doi: 10.3897/CompCytogen.v10i1.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Reports. 2016;6:28333. doi: 10.1038/srep28333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novak P, Neumann P, Pech P, Steinhaisl J, Macas J. RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013;29:792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- 33.Barghini E. The peculiar landscape of repetitive sequences in the Olive (Olea europaea L.) genome. Genome Biol. Evol. 2014;6:776–791. doi: 10.1093/gbe/evu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlek M, Gelfand Y, Plohl M, Meštrović N. Genome-wide analysis of tandem repeats in Tribolium castaneum genome reveals abundant and highly dynamic tandem repeat families with satellite DNA features in euchromatic chromosomal arms. DNA Res. 2015;22:387–401. doi: 10.1093/dnares/dsv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitkman T, et al. Next-generating sequencing reveals differentially amplified tandem repeats as major genome component of Europe’s oldest Camellia japonica. Chromosome Res. 2015;23:791–806. doi: 10.1007/s10577-015-9500-x. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn GCS. Satellite DNA transcripts have diverse biological roles in Drosophila. Heredity. 2015;115:1–2. doi: 10.1038/hdy.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu TC, Pathak S, Basen BM, Stahl GJ. Induced Robertsonian fusions and tandem translocations in mammalian cell cultures. Cytogenet. Cell. Genet. 1978;21:86–98. doi: 10.1159/000130881. [DOI] [PubMed] [Google Scholar]
- 38.Holmquist GP, Dancis B. Telomere replication, kinetochore organizers, and satellite DNA evolution. Proc. Natl. Acad. Sci. USA. 1979;76:4566–4570. doi: 10.1073/pnas.76.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer D, Downhower L. Highly repeated DNA of the baboon: organization of sequences homologous to highly repeated DNA of the African green monkey. J. Mol. Biol. 1979;134:835–842. doi: 10.1016/0022-2836(79)90488-1. [DOI] [PubMed] [Google Scholar]
- 40.Bachmann L, Venanzetti F, Sbordoni V. Tandemly repeated satellite DNA of Dolichopoda schiavazzii: a test for models on the evolution of highly repetitive DNA. J. Mol. Evol. 1996;43:135–144. doi: 10.1007/BF02337358. [DOI] [PubMed] [Google Scholar]
- 41.Stephan W, Cho S. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics. 1994;156:333–341. doi: 10.1093/genetics/136.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen S, Segal D. Extrachromosomal circular DNA in Eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet. Genome Res. 2009;124:327–338. doi: 10.1159/000218136. [DOI] [PubMed] [Google Scholar]
- 43.Inukai T. Role of transposable elements in the propagation of minisatellites in the rice genome. Mol. Genet. Genomics. 2004;271:220–227. doi: 10.1007/s00438-003-0973-5. [DOI] [PubMed] [Google Scholar]
- 44.Lal S, Oetjens M, Hannah LC. Helitrons: enigmatic abductors and mobilizers of host genome sequences. Plant Sci. 2009;176:181–186. doi: 10.1016/j.plantsci.2008.11.004. [DOI] [Google Scholar]
- 45.Macas J, Koblížková A, Navrátilová A, Neumann P. Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene. 2009;448:198–206. doi: 10.1016/j.gene.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Smýkal P, Kalendar R, Ford R, Macas J, Griga M. Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity. 2009;103:157–167. doi: 10.1038/hdy.2009.45. [DOI] [PubMed] [Google Scholar]
- 47.Thomas J, Vadnagara K, Pritham EJ. DINE-1, the highest copy number repeats in Drosophila melanogaster are non-autonomous endonuclease-encoding rolling-circle transposable elements (Helentrons) Mobile DNA. 2014;5:18. doi: 10.1186/1759-8753-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palomeque T, Muñoz-Loópez M, Carrillo JA, Lorite P. Characterization and evolutionary dynamics of a complex family of satellite DNA in the leaf beetle Chrysolina carnifex (Coleoptera, Chrysomelidae) Chromosome Res. 2005;13:795–807. doi: 10.1007/s10577-005-1013-6. [DOI] [PubMed] [Google Scholar]
- 49.Mravinac B, Ugarković D, Franjević D, Ploh lM. Long inversely oriented subunits form a complex monomer of Tribolium brevicornis satellite DNA. J. Mol. Evol. 2005;60:513–525. doi: 10.1007/s00239-004-0236-z. [DOI] [PubMed] [Google Scholar]
- 50.Koga, A. et al. Evolutionary origin of higher-order repeat structure in alpha-satellite DNA of Primate centromeres. DNA Res. 1–9 (2014). [DOI] [PMC free article] [PubMed]
- 51.Zinic SD, Ugarkovic D, Cornudella L, Plohl M. A novel interspersed type of organization of satellite DNAs in Tribolium madens heterochromatin. Chromosome Res. 2000;8:201–212. doi: 10.1023/A:1009244711527. [DOI] [PubMed] [Google Scholar]
- 52.Kuhn GCS, Sene FM, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Res. 2008;16:307–324. doi: 10.1007/s10577-007-1195-1. [DOI] [PubMed] [Google Scholar]
- 53.Reed KM, Beukeboom LW, Eickbush DG, Werren JH. Junctions between repetitive DNAs on the PSR chromosome of Nasonia vitripennis: association of palindromes with recombination. J. Mol. Evol. 1994;38:352–362. doi: 10.1007/BF00163152. [DOI] [PubMed] [Google Scholar]
- 54.Kuhn GCS, Teo CH, Schwarzacher T, Heslop-Harrison JS. Evolutionary dynamics and sites of illegitimate recombination revealed in the interspersion and sequence junctions of two nonhomologous satellite DNAs in cactophilic Drosophila species. Heredity. 2009;102:453–464. doi: 10.1038/hdy.2009.9. [DOI] [PubMed] [Google Scholar]
- 55.Alkhimova OG, et al. Diverse patterns of the tandem repeat organization in rye chromosomes. Chromosoma. 2004;113:42–52. doi: 10.1007/s00412-004-0294-4. [DOI] [PubMed] [Google Scholar]
- 56.Paço A, Adega F, Meštrović N, Plohl M, Chaves R. The puzzling character of repetitive DNA in Phodopus genomes (Cricetidae, Rodentia) Chromosome Res. 2015;23:427–440. doi: 10.1007/s10577-015-9481-9. [DOI] [PubMed] [Google Scholar]
- 57.Lepesant JMJ, et al. Chromatin structural changes around satellite repeats on the female sex chromosome in Schistosoma mansoni and their possible role in sex chromosome emergence. Genome Biol. 2012;13:R14. doi: 10.1186/gb-2012-13-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsunaga S. Junk DNA promotes sex chromosome evolution. Heredity. 2009;102:525–526. doi: 10.1038/hdy.2009.36. [DOI] [PubMed] [Google Scholar]
- 59.Deshpande N, Meller VH. Sex chromosome evolution: life, death and repetitive DNA. Fly. 2015;8:197–199. doi: 10.1080/19336934.2015.1024395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lohe AR, Hilliker AJ, Roberts PA. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frönicke L, Scherthan H. Zoo-fluorescence in situ hybridization analysis of human and lndian muntjac karyotypes (Muntiacus muntjak vaginalis) reveals satellite DNA clusters at the margins of conserved syntenic segments. Chromosome Res. 1997;5:254–261. doi: 10.1023/B:CHRO.0000032298.22346.46. [DOI] [PubMed] [Google Scholar]
- 62.Hartmann N, Scherthan H. Characterization of ancestral chromosome fusion points in the Indian muntjac deer. Chromosoma. 2004;112:213–220. doi: 10.1007/s00412-003-0262-4. [DOI] [PubMed] [Google Scholar]
- 63.Navajas-Pérez R, et al. Reduced rates of sequence evolution of Y-linked satellite DNA in Rumex (Polygonaceae) J. Mol. Evol. 2005;60:391–399. doi: 10.1007/s00239-004-0199-0. [DOI] [PubMed] [Google Scholar]
- 64.Navajas-Pérez R, et al. The origin and evolution of the variability in a Y-specific satellite-DNA of Rumex acetosa and its relatives. Gene. 2006;368:61–71. doi: 10.1016/j.gene.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Steflova P, et al. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biol. Evol. 2013;5:769–782. doi: 10.1093/gbe/evt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kejnovsky E, Hobza R, Cermak T, Kubat Z, Vyskot B. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity. 2009;102:533–541. doi: 10.1038/hdy.2009.17. [DOI] [PubMed] [Google Scholar]
- 67.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 68.Graves JAM. Evolution of vertebrate sex chromosomes and dosage compensation. Nature Rev. Genet. 2016;17:33–46. doi: 10.1038/nrg.2015.2. [DOI] [PubMed] [Google Scholar]
- 69.Sex chromosome evolution: historical insights and future perspectives. Proc. R. Soc. B. 284, 20162806 (2017). [DOI] [PMC free article] [PubMed]
- 70.Nicolas M, et al. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 2005;3:e4. doi: 10.1371/journal.pbio.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Nature. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 73.Webb GC, White MJD, Contreras N, Cheney J. Cytogenetics of the parthogenetic grasshopper Warramaba (formely Moraba) virgo and its bisexual relatives, IV, Chromosome banding studies. Chromosoma. 1978;67:309–339. doi: 10.1007/BF00285964. [DOI] [Google Scholar]
- 74.Sambrook, J. & Russel, D. W. Molecular cloning: A laboratory manual (3rd ed.) Cold Spring Harbor Laboratory Press, New York (2001).
- 75.Andrews, S. FastQC. A quality control tool for high throughput sequence datahttp://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2012).
- 76.Gordon, A. & Hannon, G. J. Fastx-toolkit. FASTQ/A short-reads pre-processing toolshttp://hannonlab.cshl.edu/fastx_toolkit (2010).
- 77.Junier, T. & Pagni, M. Dotlet: diagonal plots in a web browser, Bioinformatics 16, 178–9 (2000). [DOI] [PubMed]
- 78.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tamura K, et al. MEGA: molecular evolutionary genetics using maximun likelihood, evolutionary distance, and maximun parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers in Bioinformatics Methods and Protocols Humana Press 365–386 (1999). [DOI] [PubMed]
- 82.Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high sensitivity, fluorescence hybridization. Proc. Nat. Acad. Sci. USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cabral-de-Mello DC, Moura RC, Martins C. Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity. 2010;104:393–400. doi: 10.1038/hdy.2009.126. [DOI] [PubMed] [Google Scholar]
- 84.de Barros AV, et al. Fiber FISH reveals different patterns of high-resolution physical mapping for repetitive DNA in fish. Aquaculture. 2001;322:47–50. [Google Scholar]
- 85.Nguyen P, et al. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc. Natl. Acad. Sci. USA. 2013;110:6931–6936. doi: 10.1073/pnas.1220372110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2016).
- 87.Peterson, B. G. et al. PerformanceAnalytics package. Econometric tools for performance and risk analysishttp://r-forge.r-project.org/projects/returnanalytics/ (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


