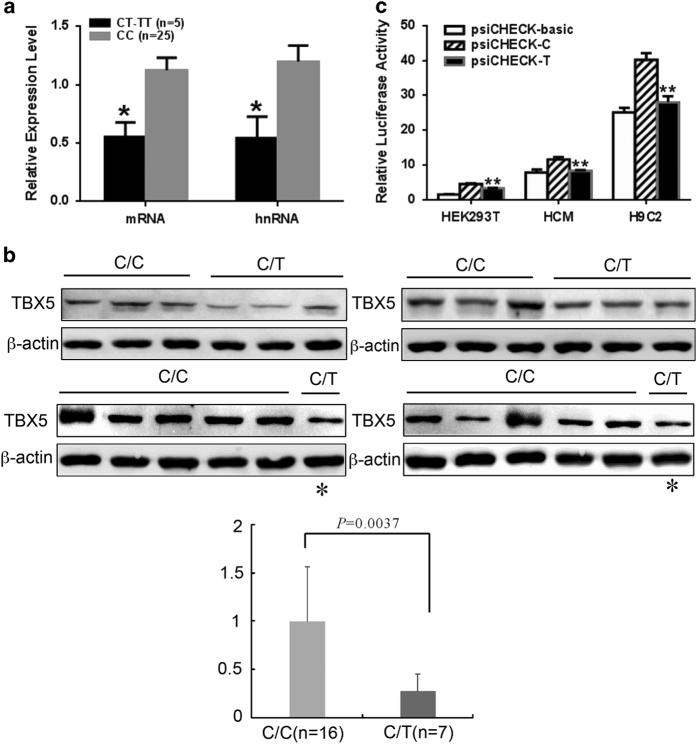
Figure 1.
The T allele of rs6489956 reduced TBX5 mRNA and protein expression levels in vivo and in vitro. (a) Quantitative real-time PCR analysis of TBX5 mRNA levels in 30 cardiac tissue samples from individuals carrying different rs6489956 C>T genotypes. The values for each genotype group were: CC=1.12±0.53 (mRNA), 1.20±0.67 (heterogenous nuclear RNA, hnRNA), CT/TT=0.55±0.27 (mRNA), 0.54±0.41 (hnRNA). All values were normalized to the levels of β-actin and represent the mean±s.d. of three independent experiments. *P<0.05. (b) Western Blot analysis in 23 cardiac tissue samples of CHDs indicated that TBX5 protein level in the CT genotype was only 27.1% that of the CC genotype (P<0.01). Considering the rare samples of the CT genotype, one of the seven tissues was used twice to compare with the CC genotype and denoted as *. (c) Luciferase expression was significantly decreased in the minor T allelic reporter compared with the major C reporter in different cells. The values in HEK 293T cells were: psiCHECK-basic=1.61±1.20, psiCHECK-C=4.60±0.39, psiCHECK-T=3.38±0.22; the values in HCM cells were: psiCHECK-basic=7.85±1.55, psiCHECK-C=11.64±2.03, psiCHECK-T=8.31±0.58; the values in H9C2 cells were: psiCHECK-basic=25.12±3.09, psiCHECK-C=40.18±5.78, psiCHECK-T =28.03±5.35. Each value represented the mean±s.d. of three independent experiments. **P<0.01.