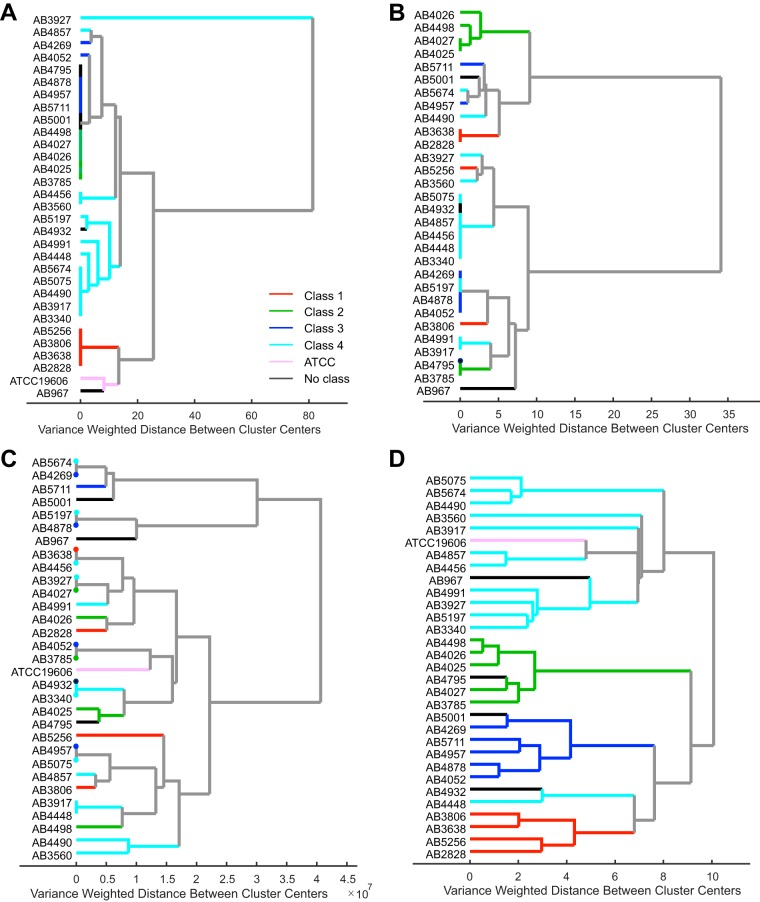
FIG 2.
Hierarchical cluster analysis (HCA) results of A. baumannii clinical isolates using DNA sequences aligned via MAFFT (multiple alignment using fast Fourier transform) and numerically coded to perform HCA using Ward's linkage and Euclidean distance (A), antimicrobial susceptibility testing data using Ward's linkage and Euclidean distance (B), preprocessed MS data performed using Ward's linkage and Euclidean distance (C), and preprocessed Raman spectroscopic data after performing principal-component analysis (D). HCA was done using Ward's linkage and Mahalanobis distance. All dendrograms are colored according to their resulting strain type (based on PFGE type, indicated by classes 1, 2, 3, and 4).