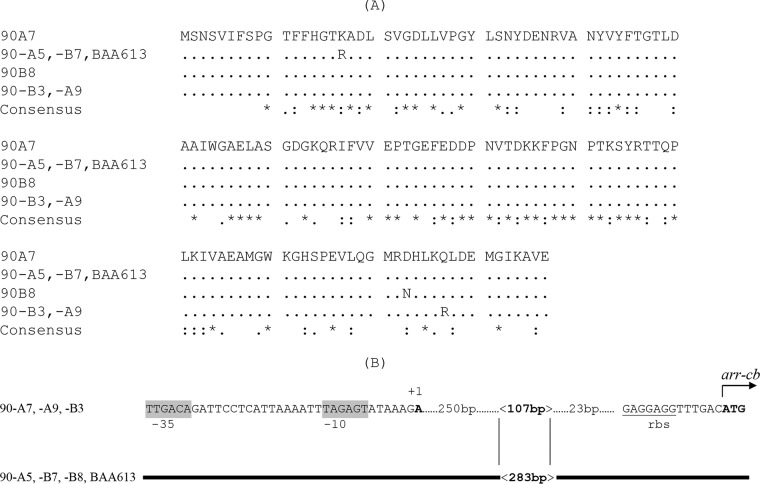
FIG 2.
Differences in Arr-cb proteins and in the regulatory regions of arr-cb in C. bolteae. (A) Deduced amino acid sequences encoded by arr-cb90A7 and its variants. Accession numbers of proteins: Arr-cb90A7, WP_002589901.1; Arr-cb90A5 and Arr-cbBAA613, WP_002569919; Arr-cb90B8, WP_002578201; Arr-cb90A9 and Arr-cb90B3, WP_002578201. The ClustalW program was used to recognize the identical amino acids encoded by arr-cb, arr-2, arr-ms, and arr-sc, which are indicated by asterisks, the highly isofunctional amino acids are indicated by two dots, and those that are isofunctional are indicated by single dots. (B) The putative regulatory region of arr-cb. The conserved sequence in C. bolteae strains is represented by the bold line. The +1 transcription initiation site identified by the 5′ RACE technique is indicated. The initiation codon of arr-cb is in bold type. The ribosome binding site is underlined. The −35 and −10 motifs, separated by 17 bp, are in gray boxes; they are conserved compared to the canonical promoter of the σ70 factor of the RNA polymerase of E. coli (TGGACA and TATAAT). In 90A5, 90B7, 90B8, and BAA613, a change from 107 to 283 bp led to a greater distance between the +1 transcription site and the initiation codon (393 to 559 bp, respectively) and acquisition of a GC-rich stem-loop (located at the beginning of the 283-bp fragment), two factors that can contribute to transcription attenuation.