ABSTRACT
There is a pressing need to identify more effective antiretroviral drugs for HIV-2 treatment. Here, we show that the investigational compound MK-8591 (4′-ethynyl-2-fluoro-2′-deoxyadenosine [EFdA]) is highly active against group A and B isolates of HIV-2; 50% effective concentrations [EC50] for HIV-2 were, on average, 4.8-fold lower than those observed for HIV-1. MK-8591 also retains potent activity against multinucleoside-resistant HIV-2 mutants (EC50 ≤ 11 nM). These data suggest that MK-8591 may have antiviral activity in HIV-2-infected individuals.
KEYWORDS: HIV-2, MK-8591, 4′-ethynyl-2-fluoro-2′-deoxyadenosine, EFdA, NRTI, antiretroviral therapy, drug resistance, HIV-1
TEXT
Antiretroviral therapy (ART) can prolong the life spans of individuals infected with human immunodeficiency virus type 1 (HIV-1) by years or even decades, but these benefits have yet to be formally demonstrated in clinical trials involving human immunodeficiency virus type 2 (HIV-2) and HIV-1/2 dually infected patients (1–3). HIV-2 is endemic in West Africa and is also prevalent in other areas with socioeconomic ties to the region (4, 5). Choices of antiretroviral (ARV) drugs for HIV-2 treatment are constrained by the intrinsic resistance of the virus to non-nucleoside reverse transcriptase inhibitors (NNRTIs) and the reduced susceptibility of HIV-2 to most HIV-1-active protease inhibitors (5–8). In addition, ART-experienced HIV-2 patients frequently harbor mutants resistant to both protease inhibitors and NRTIs (9–11), and newer ARV drugs, such as integrase strand transfer inhibitors, are not broadly available in areas where significant numbers of HIV-2-infected individuals reside. These limitations underscore the need to identify new ARV agents that exhibit potent activity against HIV-2, including drug-resistant strains of the virus.
MK-8591 (4′-ethynyl-2-fluoro-2′-deoxyadenosine [EFdA]) (Fig. 1) is an investigational nucleoside analog that inhibits HIV-1 replication with 50% effective concentrations (EC50s) in the low nanomolar-to-picomolar range (12–16). Direct comparisons show that MK-8591 is 10-fold more potent than tenofovir disoproxil fumarate (TDF) (15), 10 to100 times more potent than zidovudine (AZT) (12, 14), and 1,000 times more potent than emtricitabine (FTC) (16) against HIV-1 in culture. MK-8591 also exhibits favorable pharmacokinetic properties, including a long intracellular half-life compared to those of conventional, FDA-approved NRTIs (16–18). This property is attributable to the fluorine at the 2 position of the adenine base, which renders the compound resistant to degradation by adenosine deaminase (13, 14). MK-8591 suppresses HIV-1 and simian immunodeficiency virus (SIV) replication at clinically achievable concentrations in humanized mice and rhesus macaques, respectively (16, 19–21). In a phase 1b proof-of-concept clinical trial, a single 10-mg dose of MK-8591 demonstrated antiviral activity for 10 days in ART-naive, HIV-1-infected participants (22). The in vivo potency, pharmacokinetic profile, and physical properties of MK-8591 open the possibility for once-weekly oral dosing, and studies of long-acting formulations of MK-8591 indicate that a single parenteral administration may provide HIV-1-suppressive levels of the drug for 6 months or longer (23). Thus, MK-8591 potentially represents a new ARV with multiple modalities for preventing and treating HIV-1 infection.
FIG 1.
Chemical structures of 2′-deoxyadenosine (dA), MK-8591 (4′-ethynyl-2-fluoro-2′-deoxyadenosine [EFdA]), and BMS-986001 (2′,3′-didehydro-3′-deoxy-4′-ethynyl-thymidine; also known as festinavir, censavudine, 4′-ethynyl stavudine, 4′-ethynyl-d4T, and OBP-601).
The exceptional antiretroviral activity of MK-8591 is likely related to its unique mode of action. As with FDA-approved NRTIs, MK-8591 is converted to MK-8591-5′-triphosphate (MK-8591-TP) by cellular kinases and serves as an alternative substrate for HIV-1 reverse transcriptase (RT)-catalyzed DNA synthesis (14, 17, 24–26). However, due to its extendable 3′-hydroxyl group, MK-8591-TP is not an obligate chain terminator. Instead, incorporation of the analog results in either immediate stalling of polymerization or delayed termination after incorporation of the next complementary nucleotide (26, 27); the latter mechanism renders the newly synthesized DNA resistant to ATP- or pyrophosphate-mediated drug removal (excision) (27). In pre-steady-state kinetic assays, HIV-1 RT utilized MK-8591-TP with an efficiency comparable to or greater than observed for dATP (depending on the template sequence context), indicating that the enzyme is unable to discriminate between the two substrates (26, 27). When insertion of the analog was favored, the difference between MK-8591-TP and dATP was attributable to a lower Kd (dissociation constant) for the former (26, 27), suggesting that MK-8591-TP bound to RT with a higher affinity than dATP. This inference is supported by surface plasmon resonance data (27) and by crystallographic data from the recently solved structure of HIV-1 RT with MK-8591-TP as the incoming nucleotide (i.e., in the N site of the polymerase) (28). In this structure, the 4′-ethynyl moiety of the analog is embedded within a well-defined hydrophobic pocket in the polymerase active site. This interaction appears to stabilize MK-8591-TP in the N site complex. Additional RT structures in which MK-8591-5′-monophosphate occupies the penultimate position of the nascent strand (i.e., the P site) reveal localized distortions in the phosphate backbone that may affect nucleic acid binding and/or positioning. Taken together, the pre- and postincorporation complexes (28) reveal structural features that are consistent with MK-8591-mediated inhibition of HIV-1 RT translocation and arrest of DNA synthesis (27). To distinguish the compound from conventional chain-terminating NRTIs, MK-8591 is more accurately described as a nucleoside reverse transcriptase translocation inhibitor (NRTTI).
Although MK-8591 has been shown to potently inhibit HIV-1 and SIV in culture (12–16, 19) and in experimentally infected nonhuman primates and humanized mouse models (16, 19–21), efforts to evaluate the activity of the drug against HIV-2 are limited; a single report showed that a group B strain (HIV-2EHO) is sensitive to the drug in spreading infections of MT-4 cells (14). In addition, the ability of MK-8591 to inhibit HIV-2 mutants that are resistant to other NRTIs is unknown. In HIV-1, the M184V replacement in RT confers low-level resistance to the drug (7.5- to 15-fold increase in EC50) (13, 14), whereas the tenofovir resistance-associated change K65R confers 3- to 5-fold hypersusceptibility to MK-8591 (14, 15). However, these findings cannot be extended a priori to HIV-2, as important differences exist between the two HIV types with respect to NRTI resistance pathways and mechanisms (29–31).
We recently evaluated the antiviral activity of a 4′-ethynyl thymidine analog bearing a 2′,3′-didehydro-3′-deoxy sugar (BMS-986001; also known as festinavir or censavudine) (Fig. 1) and found that the compound had greater activity against HIV-2 than against HIV-1 in culture (32). To our knowledge, this remains the only report of an NRTI that, when tested against a diverse panel of HIV-1 and HIV-2 isolates, exhibits more-potent inhibition of HIV-2 replication. Molecular modeling suggests that the ethynyl group of BMS-986001-5′-triphosphate fits within a hydrophobic pocket in HIV-2 RT that corresponds to the aforementioned 4′ pocket of the HIV-1 enzyme (28, 32). In agreement with this model, other 4′-modified nucleosides with 2′-deoxy (14, 33) and 2′-deoxy-2′-β-fluoro (34) sugar configurations also inhibit HIV-2 replication, with EC50s in the nanomolar range. Collectively, these findings suggest that MK-8591 might inhibit a broad range of HIV-2 isolates with a potency comparable to or even better than that observed for HIV-1.
In the present study, we tested the activity of MK-8591 against a panel of HIV-1 and HIV-2 strains that were originally isolated from ART-naive individuals. We also evaluated the ability of MK-8591 to inhibit HIV-2 variants with mutations in RT that emerge in response to ART (9, 10, 35–49) and that confer resistance to various NRTIs in culture (29, 40, 50–52) and in biochemical assays with purified HIV-2 RT (30, 31). Unless otherwise specified, drug susceptibility measurements were performed using MAGIC-5A indicator cells, which are CD4+ CXCR4+ CCR5+ HeLa cells containing an HIV-inducible reporter gene (HIV long terminal repeat [LTR]–β-galactosidase). A detailed description of the assay protocol has been provided elsewhere (32). Importantly, this methodology directly quantifies antiviral activity in a single round of HIV infection (as opposed to measuring virus-induced cell death or viral protein release following multiple rounds of HIV replication), and the resultant EC50s are not affected by strain-specific differences in replication rate, infectivity, cytopathic potential, or cell-to-cell spread (32, 53, 54). The 50% cytotoxic concentration (CC50) for MK-8591 in MAGIC-5A cells was >100 nM, as determined using the CellTiter Glo assay (Promega) (32).
We initially compared the ability of MK-8591 to inhibit two prototypic isolates derived from full-length plasmid molecular clones: HIV-1NL4-3 and HIV-2ROD9. We have previously shown that these isolates exhibit comparable susceptibilities to FDA-approved NRTIs in the MAGIC-5A cell line, with EC50s that differ by 2-fold or less between the two strains (53). In the present study, we performed a total of 10 independent assay runs in which the sensitivity of HIV-1NL4-3 and HIV-2ROD9 to MK-8591 was tested head-to-head (e.g., Fig. 2A). In these experiments, the mean EC50 for HIV-1NL4-3 was 2.0 ± 0.6 nM. This result agrees with previously published measurements for HIV-1 in single-cycle assays (1.1 nM for HIV-1BH10 in MAGI cells [14], 3.2 nM for HIV-1NL101 in TZM-bl cells [15]). In contrast, within each assay run, the EC50 for HIV-2ROD9 was lower than the corresponding value for HIV-1NL4-3 by a factor of 2.3 to 9.9 (Fig. 2A and B). Following additional determinations (n = 14 total), the mean EC50 for HIV-2ROD9 was 0.42 ± 0.12 nM (Fig. 3A). Thus, MK-8591 was 4.8-fold more active against HIV-2ROD9 than against HIV-1NL4-3 in the single-cycle assay. These data are concordant with the results of spreading infection assays with CEMss cells (see reference [32] for the assay protocol), which yielded EC50s of 38 pM for HIV-2ROD9 and 120 pM for HIV-1NL4-3.
FIG 2.
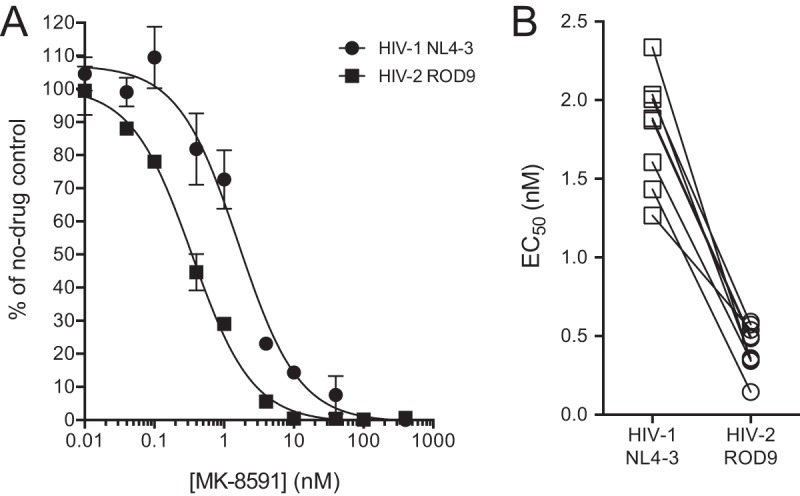
Antiviral activity of MK-8591 against HIV-1NL4-3 and HIV-2ROD9. (A) Representative data from a single run of the drug susceptibility assay in MAGIC-5A cells (single-cycle assay) (32). Data points are the percentages of β-galactosidase activity in MK-8591-treated cultures relative to that in solvent-only controls. Each point represents the mean of results from two cultures that were maintained in parallel. Error bars represent ±1 standard deviations and when not visible are smaller than the symbols. The curves were generated using a sigmoidal regression equation (GraphPad Prism 6.0 software). (B) EC50s from 10 independent runs of the single-cycle assay in which HIV-1NL4-3 and HIV-2ROD9 were tested head-to-head. Diagonal lines indicate the paired results from a single assay run.
FIG 3.
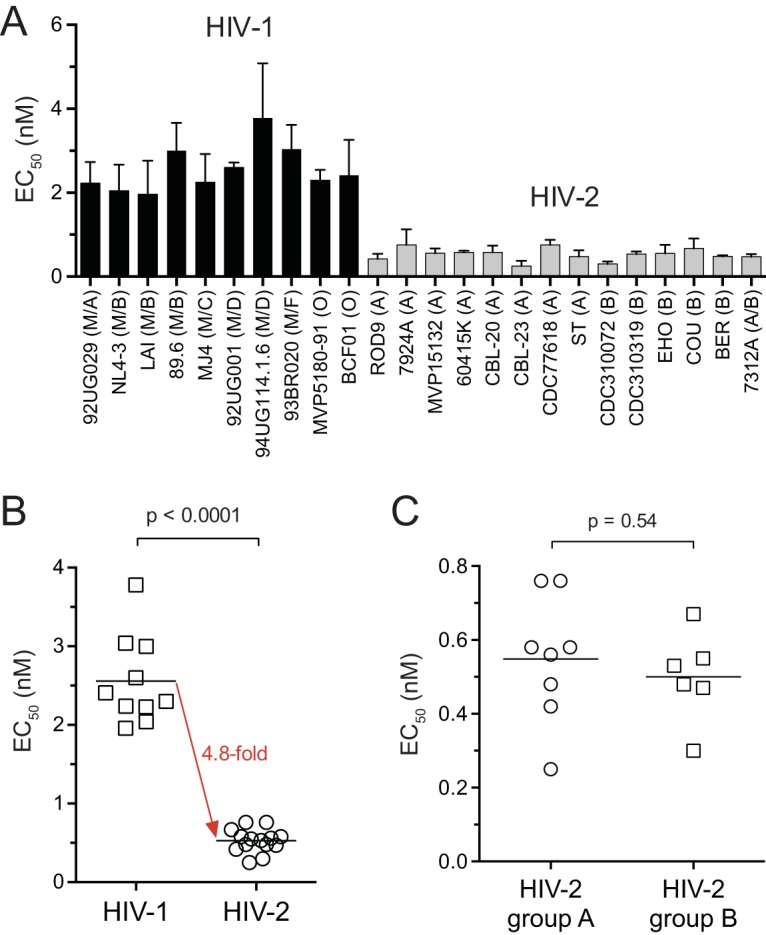
Susceptibility of HIV-1 and HIV-2 isolates from ART-naive individuals to MK-8591. (A) EC50s for 10 HIV-1 and 14 HIV-2 strains. Bars are the means of results from three or more independent single-cycle assays. Error bars are +1 standard deviation. Group/subtype assignments are shown in parentheses. The HIV-2 intergroup recombinant 7312A encodes a group B pol gene (57). (B) Summary for the isolates shown in panel A. Each point represents the mean EC50 for a single isolate. (C) Comparison of EC50s for group A and B isolates of HIV-2. In panels B and C, the horizontal lines are the means from the respective data groups; P values are the results of Welch's t tests.
Other HIV-1 and HIV-2 isolates from ART-naive individuals showed a similar trend with respect to MK-8591 susceptibility in the single-cycle assay (Fig. 3A). The mean EC50 for 10 HIV-1 strains from groups M and O was 2.6 ± 0.6 nM (range = 2.0 to 3.8 nM), whereas HIV-2 isolates from groups A and B yielded EC50s of 0.55 ± 0.17 nM and 0.50 ± 0.12 nM, respectively (range = 0.25 to 0.76 nM) (Fig. 3A and C). Overall, HIV-2 was more susceptible to MK-8591 than HIV-1 was by a difference of 4.8-fold (Fig. 3B).
To identify structural features that might explain the increased activity of MK-8591 against HIV-2, we constructed a molecular model of HIV-2 RT with MK-8591-TP bound at the polymerase active site and compared the model to the published structure of the HIV-1 RT–MK-8591-TP complex (28) (see Fig. S1 in the supplemental material). This model recapitulates important features of the HIV-1 structure, including base-pairing interactions between MK-8591-TP and the template thymidine, coordination of the triphosphate by magnesium, and the presence of a hydrophobic pocket that accommodates the 4′-ethynyl moiety of the inhibitor. However, closer inspection of the two structures suggests that subtle differences in the positioning of residues A114, Y115, F160, M184, and D185 result in a wider 4′ pocket in HIV-2 RT than in the HIV-1/MK-8591-TP complex (see Fig. S1 in the supplemental material). A similar disparity in the sizes of the 4′ pockets is evident in our models of HIV-1 and HIV-2 RT with BMS-986001-5′-triphosphate as the incoming substrate (32). Whether this variation accounts for the differential sensitivities of HIV-1 and HIV-2 to MK-8591 is unclear and will likely remain uncertain until a crystal structure of the HIV-2/MK-8591-TP complex is obtained.
Mutations that are associated with NRTI treatment can have differing effects on drug susceptibility/resistance in HIV-1 versus HIV-2, as demonstrated for thymidine analog mutations and for the Q151M replacement in RT (29–31). We therefore examined the resistance profile of MK-8591 using a panel of site-directed HIV-2ROD9 RT mutants and a recombinant clone of HIV-2 (designated 4.7a) that was derived from a patient receiving AZT, lamivudine (3TC), and ritonavir-boosted lopinavir; this clone encodes five amino acid substitutions that are associated with ART in HIV-2-infected individuals (RT changes K65R, N69S, V111I, Q151M, and M184V; see reference [32] for treatment history, clone construction, and additional details). A site-directed mutant of HIV-1NL4-3 encoding the M184V replacement was included in this analysis as an MK-8591-resistant control (14).
Relative to the parental clone, the K65R mutant of HIV-2ROD9 was hypersusceptible to MK-8591, with a mean EC50 that was 2.5-fold lower than the value for the wild-type virus; the K65R+Q151M HIV-2ROD9 mutant was also hypersusceptible to MK-8591 (2.1-fold) (Table 1). MK-8591 was fully active against Q151M HIV-2ROD9, whereas the M184V change conferred 26-fold resistance to the drug. A slightly lower level of resistance (9-fold) was observed for M184V HIV-1NL4-3 (Table 1). In contrast, in both HIV-1 and HIV-2, the M184V replacement confers >1,000-fold resistance to FTC and 3TC (50, 51, 55). Overall, the changes in MK-8591 susceptibility that resulted from single amino acid changes K65R, Q151M, and M184V in HIV-2ROD9 were similar to those reported for HIV-1 (13–15).
TABLE 1.
Susceptibilities of HIV-1 and HIV-2 RT mutants to MK-8591
| HIV clonea | Genotypeb | EC50 (nM)c | No.d | Fold changee |
|---|---|---|---|---|
| HIV-1NL4-3 | Wild type | 2.0 ± 0.62 | 10 | |
| M184V | 18 ± 6.8 | 4 | 9 | |
| HIV-2ROD9 | Wild type | 0.42 ± 0.12 | 14 | |
| K65R | 0.17 ± 0.03 | 3 | 0.4 | |
| Q151M | 0.38 ± 0.15 | 3 | 1 | |
| M184V | 11 ± 6.4 | 6 | 26 | |
| K65R+Q151M | 0.20 ± 0.06 | 3 | 0.5 | |
| K65R+M184V | 8.0 ± 2.7 | 3 | 19 | |
| Q151M+M184V | 5.9 ± 3.6 | 5 | 14 | |
| K65R+Q151M+M184V | 6.2 ± 2.0 | 3 | 15 | |
| K65R+K70R+Q151M+M184V | 7.0 ± 2.5 | 3 | 17 | |
| K65R+Y115F+Q151M+M184V | 5.3 ± 3.3 | 5 | 13 | |
| HIV-2ROD9-4.7a | K65R+N69S+V111I+Q151M+M184V | 5.4 ± 0.7 | 3 | 13 |
Viruses produced from full-length plasmids pNL4-3, pROD9, and patient-derived clone pROD9-4.7a.
Amino acid changes listed for HIV-1NL4-3 and HIV-2ROD9 were engineered by site-directed mutagenesis. Changes listed for HIV-2ROD9-4.7a were encoded by a pol gene segment that was PCR amplified from an HIV-2-infected patient (32).
EC50, 50% effective concentrations measured in the MAGIC-5A single-cycle assay. Values shown in bold are significantly different from the corresponding wild-type value (P < 0.05; analysis of variance of log10-transformed EC50s with Sidak's posttest).
Number of independent dose-response assays performed for each strain.
EC50 for the mutant divided by the EC50 for the corresponding wild-type clone (wild-type HIV-2ROD9 for the patient-derived strain).
We also observed moderate MK-8591 resistance (13- to 19-fold) for HIV-2 mutants that encode M184V in combination with one or more treatment-associated amino acid changes in RT, including those in the patient-derived clone 4.7a (Table 1). In HIV-2ROD9, the addition of up to three changes (K65R, Q151M, and either K70R or Y115F) together with M184V did not lead to higher levels of resistance than with M184V alone. It is important to note that the EC50s for all HIV-2ROD9 clones containing M184V (with or without other changes) were only 2- to 4-fold greater than the mean EC50 for HIV-1 isolates from ART-naive individuals (2.6 nM). Although we cannot exclude the possibility that other amino acid replacements in HIV-2 RT confer higher levels of MK-8591 resistance, our analysis demonstrates that MK-8591 retains substantial activity against the types of drug-resistant variants that typically emerge in NRTI-treated HIV-2 patients (9, 10, 35–49).
Lastly, we performed a series of experiments to confirm that the hypersusceptibility and resistance phenotypes observed with MK-8591 (Fig. 1 and 2 and Table 1) were not an artifact of the conditions used in the MAGIC-5A single-cycle assay. The protocol for this analysis was identical in all aspects to the one used in the above-described assays. In addition, for each strain tested in the follow-up study, the multiplicity of infection (MOI) was comparable to the MOI used in previous assay runs with MK-8591 (there was a <2-fold difference between the two experimental sets); we have previously shown that variations in MOIs of up to 32-fold have no effect on the EC50s obtained in the single-cycle assay (32). As observed in our previously published work (29, 32, 53, 56), HIV-1NL4-3 and HIV-2ROD9 were equally susceptible to AZT, stavudine [d4T], and the integrase inhibitor raltegravir, with EC50s that differed by <1.2-fold between the two strains (see Table S1 in the supplemental material). HIV-1NL4-3 and HIV-2ROD9 were also comparable to each other with respect to TDF susceptibility in this analysis (EC50s of 13 ± 1.3 nM and 6.4 ± 1.3 nM, respectively). Furthermore, HIV-2 RT mutants that were either hypersusceptible or resistant to MK-8591 (Table 1) were fully sensitive to raltegravir (maximal change in EC50 of 1.2-fold relative to the EC50 for wild-type HIV-2ROD9) (Table S2). Collectively, these data demonstrate that the MK-8591 hypersusceptibility and resistance phenotypes reported here for HIV-2 stem from intrinsic properties of the virus, rather than a systematic bias in assay setup or performance.
In summary, our findings show that MK-8591 is highly active against a broad range of HIV-2 isolates from ART-naive individuals (Fig. 3A and B). In conjunction with our earlier study of BMS-986001 (Fig. 1) (32), data from the present work provide an additional example of a nucleoside analog that exhibits greater activity against HIV-2 than against HIV-1 in a single cycle of infection. MK-8591 is also highly active against NRTI-resistant HIV-2 mutants, including variants that harbor the M184V change in RT, with EC50s of ≤11 nM (Table 1). Furthermore, the addition of K65R, Q151M, or both in combination with M184V did not lead to higher levels of MK-8591 resistance than with M184V alone. This result is significant given the frequent appearance of K65R+M184V, Q151M+M184V, and K65R+Q151M+M184V variants in HIV-2-infected patients (9, 36, 44, 48, 49). We speculate that, relative to 3TC/FTC, MK-8591 might delay the appearance of M184V in both HIV-1- and HIV-2-infected patients and, in the event of its emergence, retain substantial activity against the mutant virus. MK-8591 might also be considered for second-line or salvage therapy in patients harboring M184V variants. This could be particularly important for HIV-2 treatment, since second-line options for HIV-2 patients are restricted by both innate and emergent ARV resistance. Collectively, our findings suggest that MK-8591 may have clinical activity in HIV-2 infection.
Supplementary Material
ACKNOWLEDGMENTS
Approval for collection of data and patient samples for this study was obtained from the UW Institutional Review Board and Senegal ethics committee.
These studies were supported by grants to G.S.G. from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) (grants 1R01-AI120765 and 2R01-AI060466), the UW Center for AIDS Research (CFAR, an NIH-funded program) (grant P30 AI027757), and the UW Royalty Research Fund (grant A92723). G.S.G. has received research grants and support from the U.S. National Institutes of Health, the University of Washington, the Bill and Melinda Gates Foundation, Gilead Sciences, Alere Technologies, Merck & Co., Janssen Pharmaceutica, Cerus Corporation, and Abbott Molecular Diagnostics. V.H.W. and S.M. are recipients of undergraduate research scholarships from the Mary Gates Endowment for Students. M.S. has received support from the NIH/NIAID and France Recherche Nord & Sud Sida-hiv Hépatites (ANRS). J.A.G. is an employee of Merck & Co., Inc., Kenilworth, NJ, USA. These funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank Merck & Co., Inc., Kenilworth, NJ, USA, for provision of MK-8591.
Additional UW–Dakar HIV-2 Study Group members are as follows: Fatima Sall, Fatou Traore, Khadim Faye, Marie Pierre Sy, Bintou Diaw, Ousseynou Ndiaye, Amadou Bale Diop, Marianne Fadam Diome (Clinique des Maladies Infectieuses Ibrahima DIOP Mar, Centre Hospitalier Universitaire de Fann, Université Cheikh Anta Diop de Dakar, Dakar, Senegal); Alassane Niang, Jean Jacques Malomar, ElHadji Ibrahima Sall, Ousseynou Cisse, Ibrahima Tito Tamba, Jean Philippe Diatta, Raphael Bakhoum, Jacque Francois Sambou, Juliette Gomis (Région Médicale de Ziguinchor, Ziguinchor, Casamance, Senegal); Stephen Hawes, John Lin, Ming Chang, Robert Coombs, James Mullins, and Nancy Kiviat (University of Washington, Seattle, WA).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00744-17.
REFERENCES
- 1.Gottlieb GS, Eholie SP, Nkengasong JN, Jallow S, Rowland-Jones S, Whittle HC, Sow PS. 2008. A call for randomized controlled trials of antiretroviral therapy for HIV-2 infection in West Africa. AIDS 22:2069–2072. doi: 10.1097/QAD.0b013e32830edd44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matheron S. 2008. HIV-2 infection: a call for controlled trials. AIDS 22:2073–2074. doi: 10.1097/QAD.0b013e32830edd59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekouevi DK, Tchounga BK, Coffie PA, Tegbe J, Anderson AM, Gottlieb GS, Vitoria M, Dabis F, Eholie SP. 2014. Antiretroviral therapy response among HIV-2 infected patients: a systematic review. BMC Infect Dis 14:461. doi: 10.1186/1471-2334-14-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Silva TI, Cotten M, Rowland-Jones SL. 2008. HIV-2: the forgotten AIDS virus. Trends Microbiol 16:588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Visseaux B, Damond F, Matheron S, Descamps D, Charpentier C. 2016. HIV-2 molecular epidemiology. Infect Genet Evol 46:233–240. doi: 10.1016/j.meegid.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Menendez-Arias L, Alvarez M. 2014. Antiretroviral therapy and drug resistance in human immunodeficiency virus type 2 infection. Antiviral Res 102:70–86. doi: 10.1016/j.antiviral.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Ntemgwa ML, d'Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. 2009. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob Agents Chemother 53:3611–3619. doi: 10.1128/AAC.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menendez-Arias L, Tozser J. 2008. HIV-1 protease inhibitors: effects on HIV-2 replication and resistance. Trends Pharmacol Sci 29:42–49. doi: 10.1016/j.tips.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb GS, Badiane NM, Hawes SE, Fortes L, Toure M, Ndour CT, Starling AK, Traore F, Sall F, Wong KG, Cherne SL, Anderson DJ, Dye SA, Smith RA, Mullins JI, Kiviat NB, Sow PS, University of Washington–Dakar HIV-2 Study Group. 2009. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resource-limited West Africa. Clin Infect Dis 48:476–483. doi: 10.1086/596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charpentier C, Eholie S, Anglaret X, Bertine M, Rouzioux C, Avettand-Fenoel V, Messou E, Minga A, Damond F, Plantier JC, Dabis F, Peytavin G, Brun-Vezinet F, Ekouevi DK, IeDEA West Africa Collaboration. 2014. Genotypic resistance profiles of HIV-2-treated patients in West Africa. AIDS 28:1161–1169. doi: 10.1097/QAD.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pina-Araujo I, Guimaraes ML, Bello G, Vicente AC, Morgado MG. 2014. Profile of the HIV epidemic in Cape Verde: molecular epidemiology and drug resistance mutations among HIV-1 and HIV-2 infected patients from distinct islands of the archipelago. PLoS One 9:e96201. doi: 10.1371/journal.pone.0096201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohrui H. 2006. 2′-Deoxy-4′-C-ethynyl-2-fluoroadenosine, a nucleoside reverse transcriptase inhibitor, is highly potent against all human immunodeficiency viruses type 1 and has low toxicity. Chem Rec 6:133–143. doi: 10.1002/tcr.20078. [DOI] [PubMed] [Google Scholar]
- 13.Ohrui H, Kohgo S, Hayakawa H, Kodama E, Matsuoka M, Nakata T, Mitsuya H. 2006. 2′-Deoxy-4′-C-ethynyl-2-fluoroadenosine: a nucleoside reverse transcriptase inhibitor with highly potent activity against all HIV-1 strains, favorable toxic profiles and stability in plasma. Nucleic Acids Symp Ser (Oxf) 50:1–2. doi: 10.1093/nass/nrl001. [DOI] [PubMed] [Google Scholar]
- 14.Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int J Biochem Cell Biol 40:2410–2420. doi: 10.1016/j.biocel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Hypersusceptibility mechanism of tenofovir-resistant HIV to EFdA. Retrovirology 10:65. doi: 10.1186/1742-4690-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoddart CA, Galkina SA, Joshi P, Kosikova G, Moreno ME, Rivera JM, Sloan B, Reeve AB, Sarafianos SG, Murphey-Corb M, Parniak MA. 2015. Oral administration of the nucleoside EFdA (4′-ethynyl-2-fluoro-2′-deoxyadenosine) provides rapid suppression of HIV viremia in humanized mice and favorable pharmacokinetic properties in mice and the rhesus macaque. Antimicrob Agents Chemother 59:4190–4198. doi: 10.1128/AAC.05036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng YC, Mitsuya H. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother 51:2701–2708. doi: 10.1128/AAC.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby KA, Michailidis E, Fetterly TL, Steinbach MA, Singh K, Marchand B, Leslie MD, Hagedorn AN, Kodama EN, Marquez VE, Hughes SH, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Effects of substitutions at the 4′ and 2 positions on the bioactivity of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother 57:6254–6264. doi: 10.1128/AAC.01703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. 2012. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob Agents Chemother 56:4707–4712. doi: 10.1128/AAC.00723-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanmugasundaram U, Kovarova M, Ho PT, Schramm N, Wahl A, Parniak MA, Garcia JV. 2016. Efficient inhibition of HIV replication in the gastrointestinal and female reproductive tracts of humanized BLT mice by EFdA. PLoS One 11:e0159517. doi: 10.1371/journal.pone.0159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hattori S, Ide K, Nakata H, Harada H, Suzu S, Ashida N, Kohgo S, Hayakawa H, Mitsuya H, Okada S.. 2009. Potent activity of a nucleoside reverse transcriptase inhibitor, 4′-ethynyl-2-fluoro-2′-deoxyadenosine, against human immunodeficiency virus type 1 infection in a model using human peripheral blood mononuclear cell-transplanted NOD/SCID Janus kinase 3 knockout mice. Antimicrob Agents Chemother 53:3887–3893. doi: 10.1128/AAC.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman EJ, Schuermann D, Rudd DJ, Fox-Bosetti S, Zhang S, Robberechts M, Hueser A, Hazuda DJ, Iwamoto M, Grobler J. 2016. A single monotherapy dose of MK-8591, a novel NRTI, suppresses HIV for 10 days. Abstr Conf Retrovir Oppor Infect, abstr 437LB. [Google Scholar]
- 23.Grobler J, Friedman EJ, Barrett SE, Wood SL, Ankrom W, Filgrove KL, Lai MT, Gindy M, Iwamoto M, Hazuda DJ. 2016. Long-acting oral and parenteral dosing of MK-8591 for HIV treatment or prophylaxis. Abstr Conf Retrovir Oppor Infect, abstr 98. [Google Scholar]
- 24.Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2009. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J Biol Chem 284:35681–35691. doi: 10.1074/jbc.M109.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achuthan V, Singh K, DeStefano JJ. 2017. Physiological Mg2+ conditions significantly alter the inhibition of HIV-1 and HIV-2 reverse transcriptases by nucleoside and non-nucleoside inhibitors in vitro. Biochemistry 56:33–46. doi: 10.1021/acs.biochem.6b00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muftuoglu Y, Sohl CD, Mislak AC, Mitsuya H, Sarafianos SG, Anderson KS. 2014. Probing the molecular mechanism of action of the HIV-1 reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) using pre-steady-state kinetics. Antiviral Res 106:1–4. doi: 10.1016/j.antiviral.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michailidis E, Huber AD, Ryan EM, Ong YT, Leslie MD, Matzek KB, Singh K, Marchand B, Hagedorn AN, Kirby KA, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2014. 4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) inhibits HIV-1 reverse transcriptase with multiple mechanisms. J Biol Chem 289:24533–24548. doi: 10.1074/jbc.M114.562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2016. Structural basis of HIV inhibition by translocation-defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA). Proc Natl Acad Sci U S A 113:9274–9279. doi: 10.1073/pnas.1605223113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RA, Anderson DJ, Pyrak CL, Preston BD, Gottlieb GS. 2009. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J Infect Dis 199:1323–1326. doi: 10.1086/597802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. 2006. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog 2:e10. doi: 10.1371/journal.ppat.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer PL, Clark PK, Hughes SH. 2012. HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol 86:5885–5894. doi: 10.1128/JVI.06597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith RA, Raugi DN, Wu VH, Leong SS, Parker KM, Oakes MK, Sow PS, Ba S, Seydi M, Gottlieb GS, University of Washington–Dakar HIV-2 Study Group. 2015. The nucleoside analog BMS-986001 shows greater in vitro activity against HIV-2 than against HIV-1. Antimicrob Agents Chemother 59:7437–7446. doi: 10.1128/AAC.01326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, Mitsuya H. 2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob Agents Chemother 45:1539–1546. doi: 10.1128/AAC.45.5.1539-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang RR, Yang QH, Luo RH, Peng YM, Dai SX, Zhang XJ, Chen H, Cui XQ, Liu YJ, Huang JF, Chang JB, Zheng YT. 2014. Azvudine, a novel nucleoside reverse transcriptase inhibitor showed good drug combination features and better inhibition on drug-resistant strains than lamivudine in vitro. PLoS One 9:e105617. doi: 10.1371/journal.pone.0105617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trevino A, de Mendoza C, Caballero E, Rodriguez C, Parra P, Benito R, Cabezas T, Roc L, Aguilera A, Soriano V, HIV-2 Spanish Study Group. 2011. Drug resistance mutations in patients infected with HIV-2 living in Spain. J Antimicrob Chemother 66:1484–1488. doi: 10.1093/jac/dkr164. [DOI] [PubMed] [Google Scholar]
- 36.Descamps D, Damond F, Matheron S, Collin G, Campa P, Delarue S, Pueyo S, Chene G, Brun-Vezinet F, French ANRS HIV-2 Cohort Study Group. 2004. High frequency of selection of K65R and Q151M mutations in HIV-2 infected patients receiving nucleoside reverse transcriptase inhibitors containing regimen. J Med Virol 74:197–201. doi: 10.1002/jmv.20174. [DOI] [PubMed] [Google Scholar]
- 37.Delory T, Papot E, Rioux C, Charpentier C, Auge-Courtoi C, Michard F, Peytavin G, Descamps D, Matheron S, Yazdanpanah Y. 2016. Foscarnet, zidovudine and dolutegravir combination efficacy and tolerability for late stage HIV salvage therapy: a case-series experience. J Med Virol 88:1204–1210. doi: 10.1002/jmv.24442. [DOI] [PubMed] [Google Scholar]
- 38.Jallow S, Kaye S, Alabi A, Aveika A, Sarge-Njie R, Sabally S, Corrah T, Whittle H, Vanham G, Rowland-Jones S, Janssens W, McConkey SJ. 2006. Virological and immunological response to Combivir and emergence of drug resistance mutations in a cohort of HIV-2 patients in The Gambia. AIDS 20:1455–1458. doi: 10.1097/01.aids.0000233582.64467.8e. [DOI] [PubMed] [Google Scholar]
- 39.Colson P, Henry M, Tivoli N, Gallais H, Gastaut JA, Moreau J, Tamalet C. 2005. Polymorphism and drug-selected mutations in the reverse transcriptase gene of HIV-2 from patients living in southeastern France. J Med Virol 75:381–390. doi: 10.1002/jmv.20296. [DOI] [PubMed] [Google Scholar]
- 40.van der Ende ME, Guillon C, Boers PH, Ly TD, Gruters RA, Osterhaus AD, Schutten M. 2000. Antiviral resistance of biologic HIV-2 clones obtained from individuals on nucleoside reverse transcriptase inhibitor therapy. J Acquir Immune Defic Syndr 25:11–18. doi: 10.1097/00126334-200009010-00002. [DOI] [PubMed] [Google Scholar]
- 41.Adje-Toure CA, Cheingsong R, Garcia-Lerma JG, Eholie S, Borget MY, Bouchez JM, Otten RA, Maurice C, Sassan-Morokro M, Ekpini RE, Nolan M, Chorba T, Heneine W, Nkengasong JN. 2003. Antiretroviral therapy in HIV-2-infected patients: changes in plasma viral load, CD4+ cell counts, and drug resistance profiles of patients treated in Abidjan, Cote d'Ivoire. AIDS 17(Suppl 3):S49–S54. doi: 10.1097/00002030-200317003-00007. [DOI] [PubMed] [Google Scholar]
- 42.Brandin E, Lindborg L, Gyllensten K, Brostrom C, Hagberg L, Gisslen M, Tuvesson B, Blaxhult A, Albert J. 2003. pol gene sequence variation in Swedish HIV-2 patients failing antiretroviral therapy. AIDS Res Hum Retroviruses 19:543–550. doi: 10.1089/088922203322230905. [DOI] [PubMed] [Google Scholar]
- 43.Damond F, Matheron S, Peytavin G, Campa P, Taieb A, Collin G, Delaunay C, Chene G, Brun-Vezinet F, Descamps D. 2004. Selection of K65R mutation in HIV-2-infected patients receiving tenofovir-containing regimen. Antivir Ther 9:635–636. [PubMed] [Google Scholar]
- 44.van der Ende ME, Prins JM, Brinkman K, Keuter M, Veenstra J, Danner SA, Niesters HG, Osterhaus AD, Schutten M. 2003. Clinical, immunological and virological response to different antiretroviral regimens in a cohort of HIV-2-infected patients. AIDS 17(Suppl 3):S55–S61. doi: 10.1097/00002030-200317003-00008. [DOI] [PubMed] [Google Scholar]
- 45.Charpentier C, Camacho R, Ruelle J, Eberle J, Gurtler L, Pironti A, Sturmer M, Brun-Vezinet F, Kaiser R, Descamps D, Obermeier M. 2015. HIV-2EU-supporting standardized HIV-2 drug-resistance interpretation in Europe: an update. Clin Infect Dis 61:1346–1347. doi: 10.1093/cid/civ572. [DOI] [PubMed] [Google Scholar]
- 46.Jallow S, Alabi A, Sarge-Njie R, Peterson K, Whittle H, Corrah T, Jaye A, Cotten M, Vanham G, McConkey SJ, Rowland-Jones S, Janssens W. 2009. Virological response to highly active antiretroviral therapy in patients infected with human immunodeficiency virus type 2 (HIV-2) and in patients dually infected with HIV-1 and HIV-2 in the Gambia and emergence of drug-resistant variants. J Clin Microbiol 47:2200–2208. doi: 10.1128/JCM.01654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landman R, Damond F, Gerbe J, Brun-Vezinet F, Yeni P, Matheron S. 2009. Immunovirological and therapeutic follow-up of HIV-1/HIV-2-dually seropositive patients. AIDS 23:426–428. doi: 10.1097/QAD.0b013e328321305a. [DOI] [PubMed] [Google Scholar]
- 48.Ruelle J, Roman F, Vandenbroucke AT, Lambert C, Fransen K, Echahidi F, Pierard D, Verhofstede C, Van Laethem K, Delforge ML, Vaira D, Schmit JC, Goubau P. 2008. Transmitted drug resistance, selection of resistance mutations and moderate antiretroviral efficacy in HIV-2: analysis of the HIV-2 Belgium and Luxembourg database. BMC Infect Dis 8:21. doi: 10.1186/1471-2334-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodes B, Holguin A, Soriano V, Dourana M, Mansinho K, Antunes F, Gonzalez-Lahoz J. 2000. Emergence of drug resistance mutations in human immunodeficiency virus type 2-infected subjects undergoing antiretroviral therapy. J Clin Microbiol 38:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deuzing IP, Charpentier C, Wright DW, Matheron S, Paton J, Frentz D, van de Vijver DA, Coveney PV, Descamps D, the ANRS CO5 HIV-2 Cohort, Boucher CA, Beerens N. 2015. Mutation V111I in HIV-2 reverse transcriptase increases the fitness of the nucleoside analogue-resistant K65R and Q151M viruses. J Virol 89:833–843. doi: 10.1128/JVI.02259-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreatta K, Miller MD, White KL. 2013. HIV-2 antiviral potency and selection of drug resistance mutations by the integrase strand transfer inhibitor elvitegravir and NRTIs emtricitabine and tenofovir in vitro. J Acquir Immune Defic Syndr 62:367–374. doi: 10.1097/QAI.0b013e31827b55f1. [DOI] [PubMed] [Google Scholar]
- 52.Damond F, Collin G, Matheron S, Peytavin G, Campa P, Delarue S, Taieb A, Benard A, Chene G, Brun-Vezinet F, Descamps D, French ANRS HIV-2 Cohort. 2005. In vitro phenotypic susceptibility to nucleoside reverse transcriptase inhibitors of HIV-2 isolates with the Q151M mutation in the reverse transcriptase gene. Antivir Ther 10:861–865. [PubMed] [Google Scholar]
- 53.Smith RA, Gottlieb GS, Anderson DJ, Pyrak CL, Preston BD. 2008. Human immunodeficiency virus types 1 and 2 exhibit comparable sensitivities to zidovudine and other nucleoside analog inhibitors in vitro. Antimicrob Agents Chemother 52:329–332. doi: 10.1128/AAC.01004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith RA, Raugi DN, Pan C, Sow PS, Seydi M, Mullins JI, Gottlieb GS, University of Washington–Dakar HIV-2 Study Group. 2015. In vitro activity of dolutegravir against wild-type and integrase inhibitor-resistant HIV-2. Retrovirology 12:10. doi: 10.1186/s12977-015-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schinazi RF, Lloyd RM Jr, Nguyen MH, Cannon DL, McMillan A, Ilksoy N, Chu CK, Liotta DC, Bazmi HZ, Mellors JW. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother 37:875–881. doi: 10.1128/AAC.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith RA, Raugi DN, Kiviat NB, Hawes SE, Mullins JI, Sow PS, Gottlieb GS, University of Washington–Dakar HIV-2 Study Group. 2011. Phenotypic susceptibility of HIV-2 to raltegravir: integrase mutations Q148R and N155H confer raltegravir resistance. AIDS 25:2235–2241. doi: 10.1097/QAD.0b013e32834d8e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, Neequaye AE, Whelan TM, Ho DD, Shaw GM, Sharp PM, Hahn BH. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol 68:7433–7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


