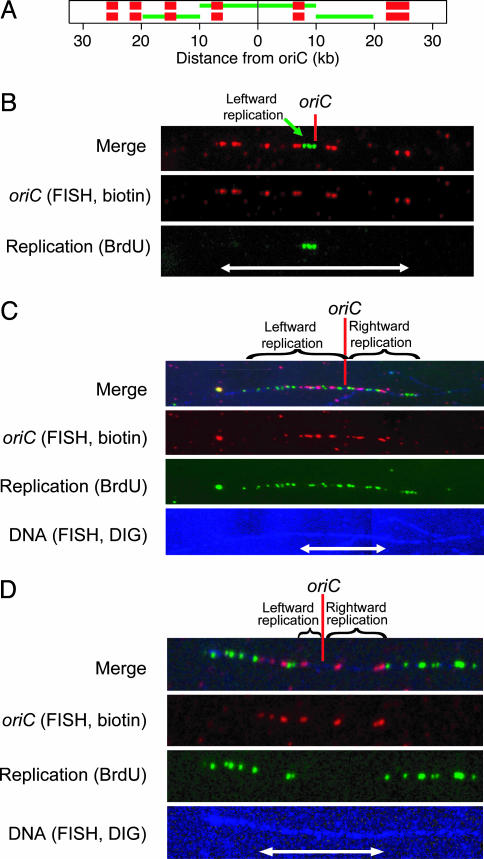
Fig. 2.
Single-molecule analysis of replication initiation. (A) Schematic diagram of expected oriC FISH and BrdUrd signals when labeling begins at (Upper) or after (Lower) initiation of replication. Four 2-kb FISH probes (red bars) are left of oriC (black dot), and one 2-kb probe and one 4-kb probe are to the right. In green is the expected BrdUrd signal for symmetric replication of 10 kb to each side. In Lower, the BrdUrd signal indicates that each replication fork was 10 kb from the origin when labeling began. (B) A combed DNA molecule showing unidirectional leftward initiation. The FISH signals (red) indicate the location and orientation of the origin. The signals, with an end-to-end distance of 16.4 μm (white arrow), have a length of 3.2 kb/μm. Replication, indicated by BrdUrd immunofluorescence (green), is visible only to the left of oriC. The replication signal is 2.3 μm or 7.3 kb long. DIG, digoxigenin. (C) A combed DNA molecule showing asymmetric, bidirectional replication. The 52-kb red FISH signal indicating oriC is 20.3 μm long. The green BrdUrd signal extends 65 kb (25.5 μm) to the left and 46 kb (18.0 μm) to the right. The genomic counterstain (blue; brightened at right for visibility) shows that the ends of the replication signal are not caused by DNA breaks. (D) A combed molecule in which replication initiated just before BrdUrd labeling. The 52-kb origin signal is 12.8 μm long (white arrow). We measured gaps of 11 kb (2.7 μm) to the left and 27 kb (6.7 μm) to the right, so the rightward fork is 16 kb ahead of its sister.