Figure 4.
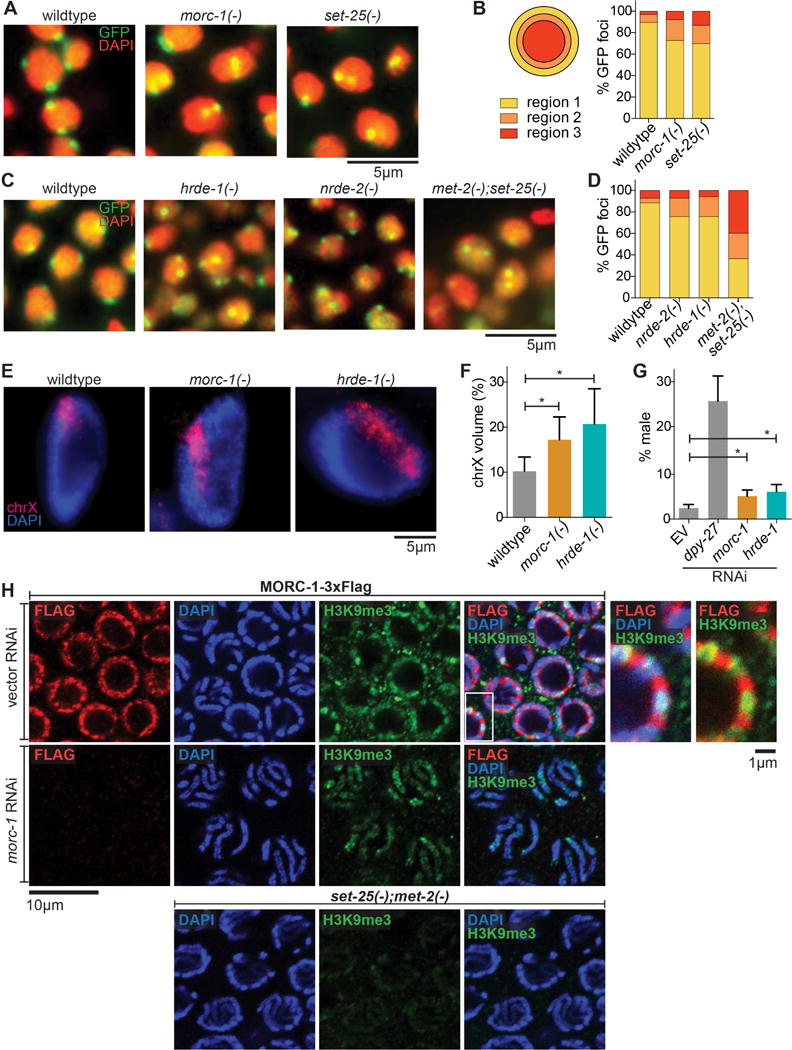
morc-1 is required for heterochromatin localization and compaction. (A) Heterochromatin localization is defective in morc-1(−) mutants. DAPI-stained embryo nuclei of the indicated genotype expressing a high-copy gwIs4[gfp-lacI::lacO] array at F4 generation at 25°C. set-25(−) is a control for mislocalization (B) Quantification is performed by determining the proportion of GFP foci in each of three concentric regions of equal volume (n≥60). (C) Localization of gwIs4 in hrde-1(−) and nrde-2(−) mutant embryos at F1 generation at 25°C using met-2(−);set-25(−) as a control for mislocalization. (D) shows quantification (n≥60) of data represented in (C). (E) Defective X chromosome compaction in intestinal nuclei of morc-1(−) and hrde-1(−) mutants. DNA FISH using a probe targeting the X chromosome and DAPI staining shows increased X chromosome volume in morc-1(−) and hrde-1(−). (F) X chromosome volume as a percentage of nuclear volume (mean±SD, n≥16) is significantly increased in morc-1(−) and hrde-1(−) intestinal nuclei (morc-1(−) vs. wildtype p=1.7×10−5, hrde-1(−) vs. wildtype p=9.6×10−6, student’s t-test). (G) RNAi against morc-1 and hrde-1 partially rescues xol-1(−);sex-1(−);him-8(−) male lethality (morc-1 vs. empty vector p=0.0053, hrde-1 vs. empty vector p=0.0018, student’s t-test). The percentage of live progeny that are male is shown as a mean of five biological replicates ± SD, n≥85. (H) Immunostaining against H3K9me3 and Flag in a strain expressing MORC-1::3xFlag on empty vector RNAi (top row). At right: high-magnification images of the region indicated by the white box with and without the DAPI channel. RNAi against morc-1 for two generations abrogates anti-Flag signal (middle row). met-2(−);set-25(−) mutants serve as a negative control for H3K9me3 staining (bottom row).
