Figure 1. Electrophilic inhibitors are targeted by nucleophilic amino acid residues.
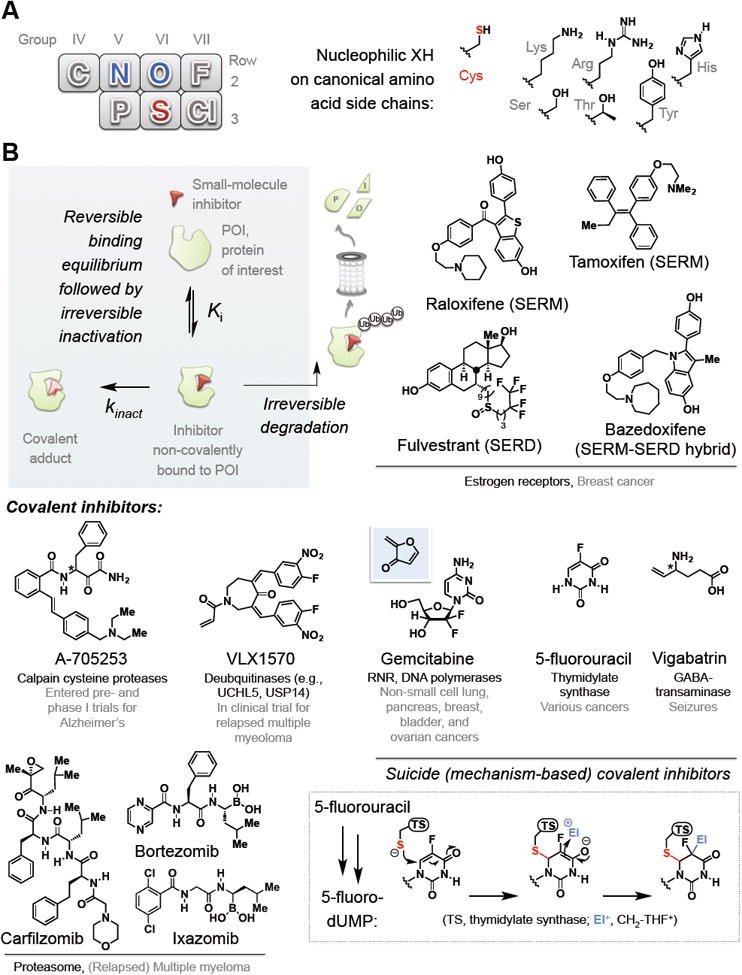
(A) (Left) Part of the periodic table showing second and third row non-metals. Blue-colored elements are components of “hard” (tend to favor carbonyl addition) nucleophilic amino acids from Row 2: sulfur (colored red) is the only primary coding residue in the third row and the only “soft” nucleophile. (Right) chemical structures of the side chains on specific residues.
(B) Clockwise from top left inset: a schematic illustration of irreversible inhibition. Top right: representative estrogen-receptors-targeting SERMs and SERDs approved for breast cancer. Middle right: examples of mechanism-based inactivators: inset at the bottom right shows currently-accepted mechanism for thymidylate synthetase (TS) inactivation by 5-FU. The active form of 5-FU (5-F-dUMP) is covalently attacked by TS, generating an enolate that traps out methylene-tetrahydrofolate (CH2-THF+) electrophile. The chemistry cannot proceed further due to the presence of the fluoride. Gemcitabine—a mechanism-based inactivator—generates the postulated reactive Michael acceptor in situ (shown above against the blue background)]. Bottom left: proteasome inhibitors present a special class of irreversible inhibition and representatives are shown. Middle left: representative covalent inhibitors of specific protein/pathway targets and disease. [Note: Stereogenic centers (*) on A-705253 and vigabatrin are undefined as they are both epimerizable in vivo].
