Figure 4. Evolutionary analyses [with emphasis on conservation of privileged cysteine(s) ] of Akt and Akt-like isoenzymes based on protein sequence data.
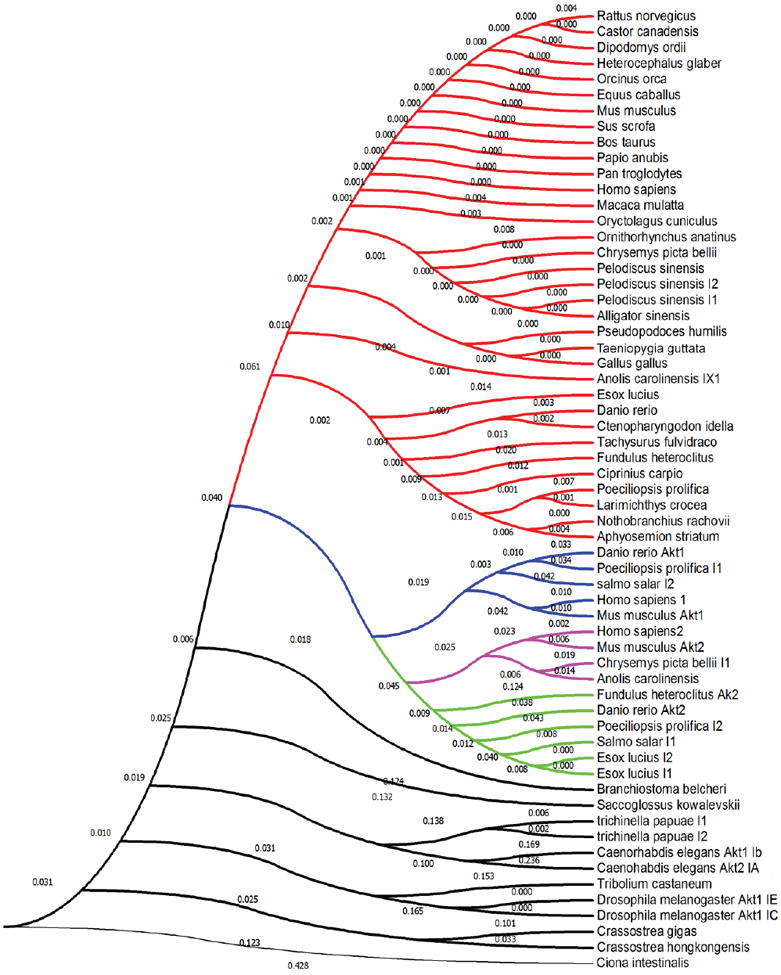
The amino acid conservation tree was constructed in MEGA-7 (Kumar, et al., 2016) using the Neighbor-Joining method (Saitou and Nei, 1987). The analysis involved 61 Akt sequences (whole-protein amino acid sequence) spanning Akt1–3 from indicated species, with the emphasis on Akt3. Red indicates Akt3-like proteins all of which contain the privileged cysteine analogous to C119. Blue indicates Akt1-like proteins that bear no cysteine(s) analogous to C119 and/or C124. Magenta indicates Akt2-like proteins with ROS-sensor cysteine (C124). Green indicates Akt2-like proteins that lack the ROS-sensor cysteine (C124). Black indicates Akt-like proteins from lower eukaryotes. Note: No protein aligning with Akt3 but lacking C119 was found in this set (see Fig. 3B for sequence logo). (The optimal tree with the sum of branch length = 2.99180282 is shown). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site.
