Figure 2.
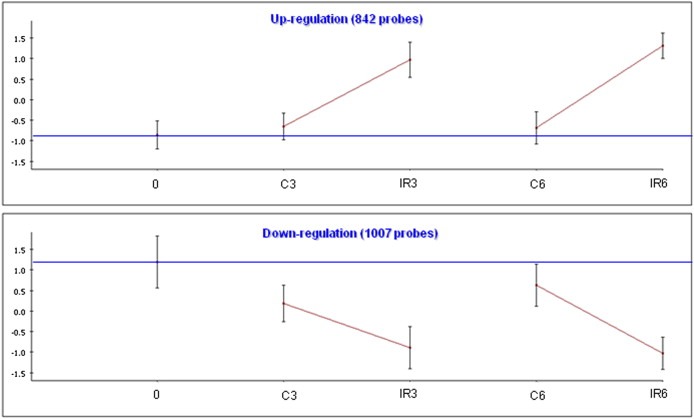
IR‐responsive gene clusters in TK6 cells. We used the clustering algorithm CLICK to divide the set of responding genes in each cell line into clusters, each of which represents a set of genes with similar expression patterns. The total number of genes in each cluster is indicated. For each cluster, the graph shows the mean expression pattern of all its genes. Error bars represent one S.D. Prior to clustering, gene expression levels were standardized to mean = 0 and SD = 1; the y‐axis corresponds to the standardized levels. The horizontal blue line represents the expression level in the untreated t0 sample (‘basal expression level’). The x‐axis corresponds to the examined conditions: C: untreated control. IR: irradiated cells. Post‐irradiation time points are indicated (0, 3 or 6 h).
