Abstract
The clinical benefit of anthracyclines has been connected to HER2 status, TOP2A status and centromere 17 copy numbers (CEN‐17). Data from a clinical trial randomizing patients to anthracyclines was used to assess whether the number of CEN‐17 in breast cancers may predict incremental responsiveness to anthracyclines besides what is obtained when used relatively to TOP2A and HER2. As cut sections of paraffin‐embedded tissue are prone to truncation of nuclei, strict definition of ploidy levels is lacking. We therefore used normal breast tissue to assist define ploidy levels in cut sections. Fluorescence in situ hybridization (FISH) with centromere 17 (CEN‐17) and TOP2A was performed on 120 normal breast specimens. The diploid CEN‐17 copy number was reduced from the expected two signals in whole nuclei to an average of 1.68 signals per nucleus in cut sections of normal breast. Ploidy levels determined in normal breast were applied to data on 767 patients with known HER2 and TOP2A status randomized to anthracyclines in the DBCG 89D trial. CEN‐17 ploidy levels were in cut sections from the 767 breast cancer patients established as: Haploid: ≤1.25 (10%), diploid: 1.26–2.09 (60%), triploid: 2.10–2.93 (21%), tetraploid: 2.94–3.77 (5%) or higher ploidy: ≥3.78 (4%). Amplification of HER2 and deletion of TOP2A were frequently observed in tumors with a high ploidy level. In univariate analyses increasing ploidy was associated with decreased disease‐free survival (DFS) (P=0.0001) and overall survival (OS) (P<0.0001). However, in multivariate analysis CEN‐17 was not established as an independent prognostic factor and was neither a statistically significant predictor of benefit from CEF (Cyclophosphamide/Epirubicin/5‐Fluorouracil) compared to CMF (Cyclophosphamide/Methotrexate/5‐Fluorouracil) (PInteraction 0.39 for DFS and 0.67 for OS). In conclusion, CEN‐17 levels do not independently from TOP2A/CEN‐17 ratio identify breast cancer patients who achieve an incremental benefit from adjuvant anthracyclines.
Keywords: Centromere 17, <i>HER2</i> , <i>TOP2A</i> , Ploidy level, Aneuploidy, Polysomy
Highlights
Overall, breast cancer patients benefit from treatment with anthracyclines.
Predictive biomarkers are needed for stratifying patients to anthracyclines.
TOP2A, HER2 or centromere 17 status may serve as biomarkers.
TOP2A but not HER2 has predictive value in the DBCG 89D trial.
CEN‐17 status does not independently from TOP2A predict anthracycline benefit.
1. Introduction
In patients with primary breast cancer, amplification of HER2 is not only a well established prognostic factor (Slamon et al., 1987; Tandon et al., 1989) and a requisite for the effect of HER2 targeted therapy, but may also predict an incremental benefit from anthracycline containing chemotherapy (Dhesy‐Thind et al., 2008; Gennari et al., 2008). Further, benefit from poisoning topoisomerase II‐α by anthracyclines (Guano et al., 1999) has in several randomized trials been associated with copy number changes in TOP2A (Knoop et al., 2005; O'Malley et al., 2009; Press et al., 2011), but not consistently (Bartlett et al., 2008b). A recent meta‐analysis has identified TOP2A aberrations (combined amplifications and deletions) as the most powerful predictor of anthracycline benefit (Leo et al., 2011).
Fluorescence in situ hybridization (FISH) is the method of choice for detection of gene copy number variation of medium‐sized DNA segments, like amplification and deletion of single genes i.e. HER2 and TOP2A. It is routinely used on cut sections of formalin‐fixed paraffin‐embedded (FFPE) tissue, but the diameter of cancer cell nuclei is generally larger than the thickness of cut sections of FFPE tissues leading to truncated nuclei. To avoid misclassification following chromosome complement doubling, i.e. polyploidization, in proliferating tissue with a high mitotic index a reference probe is included, preferentially centromere 17 (CEN‐17) concerning HER2 and TOP2A FISH. Comparative genomic hybridization (CGH), loss of heterozygosity (LOH) and molecular genetics data, taken together, show that chromosome 17 is rearranged in breast tumors (Marchio et al., 2009; Troxell et al., 2006; Yeh et al., 2009). Amplification of the centromeric region is more frequently in HER2 amplified tumors (19%) than in HER2 normal tumors (1%) (Staaf et al., 2010). Short and long arms differ in the type of events they harbor. Chromosome 17p is principally involved in losses whereas 17q shows complex combination of overlapping gains and losses (Orsetti et al., 2004; Staaf et al., 2010), indicating that the centromere of chromosome 17 may not be an ideal reference probe as measurement of the ploidy level of the tumor and caution should be taken not to overinterpret the centromere 17 copy numbers (Viale, 2009).
The inclusion of centromere 17 as reference probe has lead to debate and investigation of the importance of CEN‐17 copy number changes (reviewed by (Reinholz et al., 2009)), but the frequency of CEN‐17 copy number changes has not yet been established due to lack of agreement on the definition of polysomy. Recent studies (Marchio et al., 2009; Moelans et al., 2010; Staaf et al., 2010; Yeh et al., 2009) have shown that gain of whole chromosome 17, often referred to as polysomy 17, is rare. Nevertheless has polysomy 17 extensively been used as a proxy for centromere 17 copy number changes and we will in the absence of a generally accepted definition (Wolff et al., 2007) use this term as well.
Breast cancers are most often aneuploid, and hold a total number of chromosomes that is not a multiple of the haploid chromosome number. In contrast, polyploidy refers to a numerical change in the entire set of chromosomes. This distinction is important, but requires methodologies that are able to detect overall changes in chromosome numbers e.g. classical cytogenetic karyotyping or multicolor FISH on metaphase spreads. Fluorescence in situ hybridization used on interphase nuclei is however not capable of making this distinction (Wolman, 1994), and gene or centromere copy numbers do not provide evidence to distinguish aneuploidy from polyploidy. A complicating factor is that an aneuploid cell can even undergo polyploidization. Interphase FISH is well suited for measurement of gene and centromere copy numbers, but cannot discriminate between whole chromosome gain and single locus gain (Wolman, 1994).
We used normal breast tissue from 120 women without a history of breast cancer to define the normal ploidy levels of CEN‐17 in the truncated nuclei of cut tissue sections according to a classical cytogenetic definition and applied this definition to the CEN‐17 copy numbers obtained in a retrospective correlative science study of a randomized trial (Ejlertsen et al., 2007; Knoop et al., 2005; Nielsen et al., 2008) where HER2 and TOP2A status was known. The prognostic and predictive influence of altered copy numbers on disease‐free survival and overall survival was investigated.
2. Material and methods
2.1. Normal breast tissue
From the archives of Department of Pathology at Herlev University Hospital, 120 tissue samples from normal breasts were collected consecutively and retrospectively from breast reduction surgeries where no malignancy was identified. Tissue microarrays (TMAs) were constructed using Advanced Tissue Arrayer, ATA‐100 (Chemicon International, Temecula, CA, USA) from formalin‐fixed, paraffin‐embedded normal breast tissue. A pathologist (EB) identified areas of normal breast glands suitable for TMA construction. Duplicate 2.0mm tissue cores from each donor block were re‐embedded into recipient paraffin block containing 40 tissue cores.
2.2. Tumor tissue from invasive breast cancer
The design of the original clinical trial and the biological sub‐study has been described previously in detail (Ejlertsen et al., 2007; Knoop et al., 2005). DBCG trial 89D was an open‐label randomized, phase III, trial comparing CEF (Cyclophosphamide/Epirubicin/5‐Fluorouracil) against CMF (Cyclophosphamide/Methotrexate/5‐Fluorouracil). The Danish Breast Cancer Cooperative Group prepared the original protocol (DBCG trial 89D) as well as the biomarker sub‐study protocol (DBCG 89D/TOP2A) and The Danish National Committee on Biomedical Research Ethics approved both before activation. The biomarker analysis was preformed according to REMARK (McShane et al., 2005).
From January 1990 to January 1998, 1224 patients were randomized in DBCG trial 89D and 980 of these were recruited in Denmark. Archival paraffin‐embedded tissue blocks from 806 Danish patients enrolled in the trial were collected between September 2001 and August 2002 from the study sites and stored centrally. Presence of invasive carcinoma was confirmed on all tissue blocks. Data from the TOP2A FISH analysis were available on 773 samples. The survival analysis included 767 patients as 6 patients had unknown covariables (Nielsen et al., 2008). Paired TOP2A and HER2 FISH results were available from 637 of these patients. The main reason for the reduced number of HER2 data was a decision by the study group not to analyze the remaining cases with HercepTest score 0 after the first 100 samples were analyzed without the finding of a single amplification.
2.3. FISH analysis
TOP2A and HER2 gene aberrations were identified by FISH (K5333 TOP2A FISH pharmDx™ Kit and K5331 HER2 FISH pharmDx™ Kit, Dako, Denmark) according to the manufactures instructions. Further details are described elsewhere (Knoop et al., 2005; Nielsen et al., 2004; Olsen et al., 2004). Briefly the method consists of pretreatment for 10min at 95–99°C in pretreatment solution followed by pepsin treatment for 10min at room temperature. After denaturation at 82°C for 5min and overnight hybridization at 45°C, excess of probe was removed by stringent wash at 65°C for 10min. The TOP2A probe covers a 229kb segment containing the TOP2A gene and 2 flanking genes on each side. The probe is located 679kb from the HER2 gene. The HER2 probe covers a 218kb segment containing 5 flanking genes. TOP2A and HER2 ratio was calculated as the number of signals for the red gene probe divided by the number of signals for the green centromere 17 reference probe (CEN‐17) targeting the repetitive α‐satellite locus on 17p11.1–q11.1 (D17Z1). Cases were scored as HER2 or TOP2A FISH amplified when the ratio was ≥2.0. TOP2A cases were scored as deleted when the ratio was <0.8. HER2 deletion was also recorded using the same cutoff. Scoring of signals in breast cancer cells has been described earlier (Olsen et al., 2004). Ploidy level is based on average counts of centromere 17 in each sample. Ploidy levels were defined according to standard cytogenetic nomenclature (ISCN, 2009).
2.4. Statistical analysis
Overall survival (OS) was defined as time elapsed from randomization until death or censoring. Follow‐up for OS was achieved by linkage to the Danish Central Population Registry, with complete follow‐up as of December 1st, 2010, except for eight of the patients who emigrated (between 3.2 and 18.1 years after randomization). Disease‐free survival (DFS) was defined as the time elapsed from randomized assignment until invasive breast cancer recurrence, new invasive breast cancer, second primary non‐breast cancer, or death as first event irrespective of cause. DFS and OS were analyzed univariate using Kaplan–Meier estimates, the log rank test and the Cox proportional hazards model. The Cox proportional hazards model was also applied for multivariate analysis, based on the model developed previously for the same patient material (Knoop et al., 2005). The multivariate model included ploidy, TOP2A, HER2, histological type and grade, tumor size, number of positive lymph nodes, menopausal status, hormone receptor status and treatment with CMF or CEF. Age and menopausal status were closely associated, therefore age was not included. The Cox proportional hazards model was adjusted according to the results of the goodness‐of‐fit procedures. Hormone receptor status and histological type and grade violated the proportional hazards assumption and were included as stratification variables. The hazards ratio (HR), the 95% confidence interval (CI) and the P‐value of the Wald test were estimated for each covariate and interaction term in Cox model. Correlations between ploidy and clinical and pathological variables including HER2 and TOP2A status were tested (excluded unknowns) by χ 2‐test. Correlation between CEN‐17 and the two assays was analyzed by Pearson's correlation coefficient. P‐values are two‐tailed. Statistical analyses were done with the SAS 9·1 program package (SAS Institute, Cary, NC, USA).
3. Results
3.1. Normal breast tissue
Formalin‐fixed, paraffin‐embedded (FFPE) normal breast tissue blocks were available from 120 women who underwent breast reduction surgery and had no history of breast cancer. By TOP2A FISH an average number of 1.68 green centromere 17 (CEN‐17) signals and 1.81 red TOP2A signals were counted (Figure 1A) corresponding to a TOP2A/CEN‐17 ratio of 1.08 with a standard deviation of 0.06 (Figure 1B). The haploid number (n) of CEN‐17 signals in truncated nuclei was calculated as 0.84 (Table 1). Consequently, the diploid range (2n) was defined as 1.26 (1.68−(0.5×0.84)) to 2.09 (1.68+(0.5×0.84)), the triploid range (3n) as 2.10–2.93, the tetraploid range (4n) as 2.94–3.77 and higher ploidy level (4n+) as ≥3.78/nuclei. The term “highploid” will be used to define this group, as the FISH method cannot discriminate between polyploidy and aneuploidy. Monosomy (haploid number=n) was defined as <1.26 mean signals per nuclei. This definition places multiplex of the haploid CEN‐17 number in the middle of each defined range.
Figure 1.
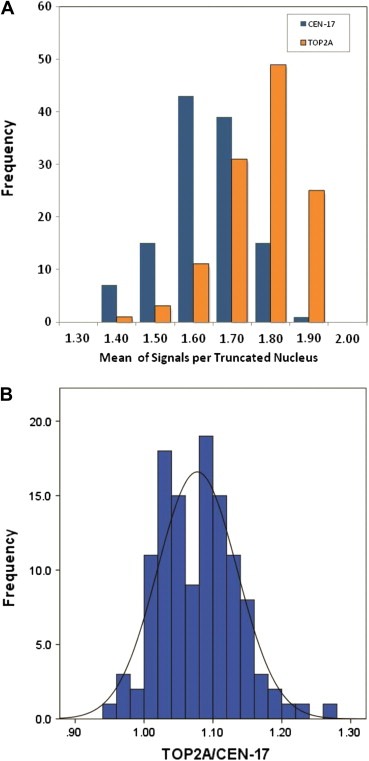
Distribution of mean number of TOP2A and centromere 17 signals in 60 nuclei/sample from a total of 120 samples of normal breast (A). Distribution of TOP2A/CEN‐17 ratios in 120 samples of normal breast (B).
Table 1.
Theoretical ploidy levels and ploidy ranges based on the average of 1.68 centromere 17 in truncated nuclei (CEN‐17 per cell) measured in 120 normal breast tissues
| Ploidy level | Centromere 17 copy number | ||
|---|---|---|---|
| Whole nuclei | Truncated nuclei | ||
| Average | Range | ||
| Monosomy/monosomic/haploid | 1 | 0.84 | <1.26 |
| Disomy/disomic/diploid | 2 | 1.68 | [1.26–2.10) |
| Trisomy/trisomic/triploid | 3 | 2.52 | [2.10–2.94) |
| Tetrasomy/tetrasomic/tetraploid | 4 | 3.36 | [2.94–3.78) |
| High ploidy level/“highploid” | >5 | Times 0.84 | ≥3.78 |
3.2. Invasive breast cancer
Information on HER2 status, TOP2A status and other covariates was available on 767 of the 980 Danish participants in the DBCG 89D trial. Paired average numbers of centromere 17 (CEN‐17) signals per nucleus in HER2 and TOP2A assays were available from 637 of these patients. A highly statistically significant correlation of the mean CEN‐17 signals per cell was determined between the two assays (Pearson correlation coefficient 0.82; P<0.0001). The mean difference in CEN‐17 counts per cell in the HER2 and TOP2A assay was 0.06. In the subsequent analyses the average of CEN‐17 signals in the two assays was used or if unavailable the CEN‐17 results from the TOP2A assay were used.
The correlation between ploidy level and other covariates for the 767 patients included in the statistical analysis is listed in Table 2. When the ploidy levels were standardized according to our findings in normal breast tissue, 80 (10%) breast cancer patients had a tumor with monosomy of CEN‐17, 458 (60%) diploid, 161 (21%) triploid, 38 (5%) tetraploid, and 30 (4%) “highploid” tumors. Increasing CEN‐17 copy number was significantly associated with the presence of HER2 amplification and TOP2A deletion (each P<0.0001). A significant association was also found for being postmenopausal (P=0.04), node positive (P=0.01) and hormone receptor negative (P=0.04), while tumor size and tumor grade are independent of CEN‐17 copy number as measurement of the ploidy level.
Table 2.
Correlation between ploidy level and other patient characteristics for 767 patients participating in the DBCG 89D trial.
| Haploid | Diploid | Triploid | Tetraploid | Highploid | Total | P‐value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | (%) | N | (%) | N | (%) | N | (%) | N | (%) | N | ||
| All patients | 80 | (10.4) | 458 | (59.7) | 161 | (21.0) | 38 | (5.0) | 30 | (3.9) | 767 | |
| Menopausal status | ||||||||||||
| Premenopausal | 56 | (70.0) | 337 | (73.6) | 101 | (62.7) | 24 | (63.2) | 14 | (46.7) | 532 | 0.004 |
| Postmenopausal | 24 | (30.0) | 121 | (26.4) | 60 | (37.3) | 14 | (36.8) | 16 | (53.3) | 235 | |
| Tumor size | ||||||||||||
| 0–20mm | 36 | (45.0) | 192 | (41.9) | 64 | (39.8) | 16 | (42.1) | 7 | (23.3) | 315 | 0.24 |
| 21–50mm | 39 | (48.8) | 229 | (50.0) | 83 | (51.6) | 16 | (42.1) | 22 | (73.3) | 389 | |
| ≥51mm | 5 | (6.3) | 37 | (8.1) | 14 | (8.7) | 6 | (15.8) | 1 | (3.3) | 63 | |
| No. of pos. nodes | ||||||||||||
| Negative | 28 | (35.0) | 189 | (41.3) | 52 | (32.3) | 10 | (26.3) | 3 | (10.0) | 282 | 0.01 |
| 1–3 pos. | 22 | (27.5) | 139 | (30.3) | 54 | (33.5) | 17 | (44.7) | 12 | (40.0) | 244 | |
| ≥4 pos. | 30 | (37.5) | 130 | (28.4) | 55 | (34.2) | 11 | (28.9) | 15 | (50.0) | 241 | |
| TOP2A FISH | ||||||||||||
| Deleted | 1 | (1.3) | 23 | (5.0) | 29 | (18.0) | 14 | (36.8) | 19 | (63.3) | 86 | <0.0001 |
| Normal | 67 | (83.8) | 385 | (84.1) | 107 | (66.5) | 21 | (55.3) | 9 | (30.0) | 589 | |
| Amplified | 12 | (15.0) | 50 | (10.9) | 25 | (15.5) | 3 | (7.9) | 2 | (6.7) | 92 | |
| HER2 FISH | ||||||||||||
| Deleted | 6 | (1.3) | 7 | (4.3) | 2 | (5.3) | 1 | (3.3) | 16 | <0.0001 | ||
| Normal | 33 | (41.3) | 272 | (59.4) | 72 | (44.7) | 14 | (36.8) | 10 | (33.3) | 401 | |
| Amplified | 23 | (28.8) | 111 | (24.2) | 60 | (37.3) | 19 | (50.0) | 19 | (63.3) | 232 | |
| Unknown | 24 | (30.0) | 69 | (15.1) | 22 | (13.7) | 3 | (7.9) | 118 | |||
| HercepTest score | ||||||||||||
| 0=neg | 36 | (45.0) | 128 | (27.9) | 35 | (21.7) | 7 | (18.4) | 206 | <0.0001 | ||
| 1+ | 18 | (22.5) | 187 | (40.8) | 45 | (28.0) | 4 | (10.5) | 2 | (6.7) | 256 | |
| 2+ | 4 | (5.0) | 44 | (9.6) | 22 | (13.7) | 5 | (13.2) | 3 | (10.0) | 78 | |
| 3+ | 22 | (27.5) | 99 | (21.6) | 59 | (36.6) | 22 | (57.9) | 25 | (83.3) | 227 | |
| Treatment | ||||||||||||
| CMF | 40 | (50.0) | 246 | (53.7) | 95 | (59.0) | 20 | (52.6) | 17 | (56.7) | 418 | 0.70 |
| CEF | 40 | (50.0) | 212 | (46.3) | 66 | (41.0) | 18 | (47.4) | 13 | (43.3) | 349 | |
| Hormone receptor | ||||||||||||
| Negative | 54 | (67.5) | 298 | (65.1) | 111 | (68.9) | 26 | (68.4) | 27 | (90.0) | 516 | 0.04 |
| Positive | 19 | (23.8) | 132 | (28.8) | 40 | (24.8) | 10 | (26.3) | 1 | (3.3) | 202 | |
| Unknown | 7 | (8.8) | 28 | (6.1) | 10 | (6.2) | 2 | (5.3) | 2 | (6.7) | 49 | |
| Tumor grade | ||||||||||||
| Grade I | 4 | (5.5) | 33 | (7.7) | 12 | (7.8) | 2 | (5.7) | 2 | (6.7) | 53 | 0.98 |
| Grade II | 39 | (48.8) | 207 | (45.2) | 80 | (49.7) | 19 | (50.0) | 15 | (50.0) | 360 | |
| Grade III | 30 | (37.5) | 190 | (41.5) | 61 | (37.9) | 14 | (36.8) | 13 | (43.3) | 308 | |
| Non‐ductal | 7 | (8.8) | 28 | (6.1) | 8 | (5.0) | 3 | (7.9) | 0 | (0.0) | 46 | |
3.3. Association of CEN‐17 alterations with outcome
This analysis was conducted 13 years after trial commencement. Estimated median potential follow‐up for OS was 16 years and 11 months and there were 368 disease‐free survival (DFS) events and 377 deaths among the 767 patients assessable for CEN‐17. Increasing CEN‐17 levels were statistically significant associated with DFS events (Figure 2A; P=0.0001) and deaths (Figure 2B; P<0.0001) in the univariate analysis.
Figure 2.
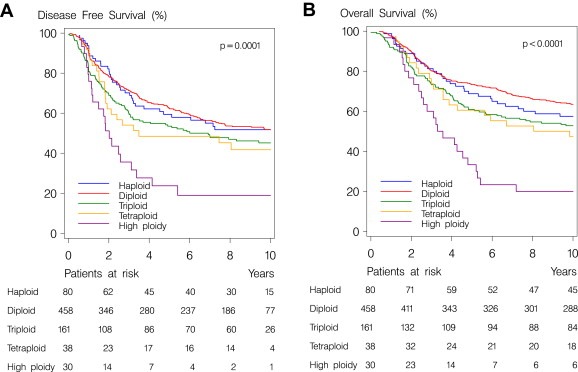
Kaplan–Meyer plot of disease‐free survival (A) and overall survival (B) of 767 breast cancers patients according to the ploidy level of the tumors.
After adjustment in a multivariate Cox model for menopausal status, tumor size, positive nodes, ER status, malignancy grade and treatment, a statistically significant heterogeneity in outcome could not be shown within haploid and diploid (P=0.24 for DFS events and P=0.20 for deaths) and in the following analysis we combined these two subgroups (CEN‐17 normal). Likewise, those with tri‐, tetra‐, and “highploid” tumors were combined (CEN‐17 altered) due to absence of heterogeneity in outcome (P=0.32 for DFS events and P=0.46 for deaths). Patients whose tumors showed CEN‐17 alterations had statistically significant higher risk of a DFS event (HR=1.36; 95% CI 1.09–1.69, P=0.006) and death (HR=1.42; 95% CI 1.14–1.96, P=0.002). However, no significant association of CEN‐17 status was observed with DFS or OS when HER2 (P=0.35 and P=0.07, respectively) and TOP2A (P=0.35 and P=0.14, respectively) status were added to the model.
3.4. Association of CEN‐17 alterations with therapy
In a multivariate Cox regression analysis, we further examined heterogeneity of treatment effect according to HER2, TOP2A and CEN‐17 status. As previously reported, for both DFS and OS, the improvement if treated with CEF compared to CMF was similar in HER‐positive and HER2‐negative tumors with no significant interaction between HER2 status and treatment effect (Figure 3) (Knoop et al., 2005; Nielsen et al., 2008). Significant interaction between TOP2A status and treatment effect was conserved for DFS (P=0.01) but only borderline for OS (P=0.06). For both DFS (Figure 3A) and OS (Figure 3B) the improvement when treated with CEF compared to CMF, seemed to be equal in patients with CEN‐17 altered and CEN‐17 normal tumors (P Interaction=0.39 for OS and P Interaction=0.67 for DFS), and broadly similar results came out whether CEN‐17 was split in two or five subgroups (Figure 3A and B). Among patients classified as having HER2 and TOP2A normal tumors (N=483) the treatment effect of CEF compared to CMF was borderline statistically significant improved for those with CEN‐17 normal status compared to the CEN‐17 altered group of patients.
Figure 3.
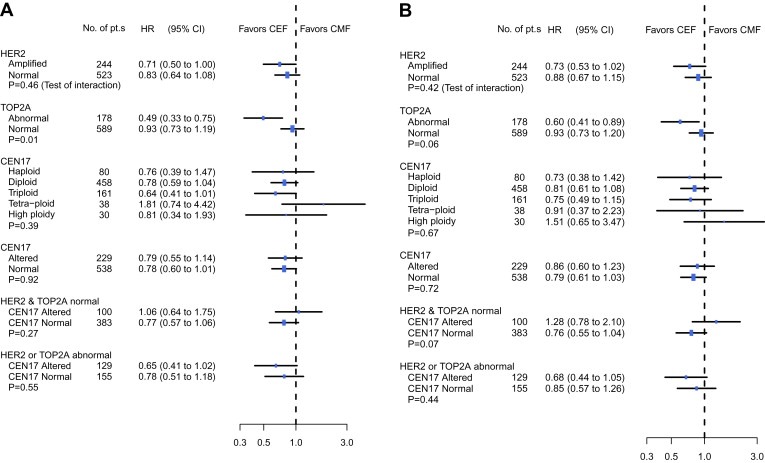
Forest plots illustrating adjusted hazard ratio estimates of treatment effect for disease‐free survival (A) and overall survival (B). P‐values refer to Wald test of heterogeneity of treatment effect and listed subgroups.
4. Discussion
In this study, we characterized centromere 17 (CEN‐17) ploidy levels in the truncated nuclei of cut sections of normal breast tissue. This classification was then easily applied to CEN‐17 results obtained by HER2 and TOP2A FISH in a retrospective correlative science study of the DBCG 89D trial. Only 30 (3.9%) of the 767 patients had tumors with very high ploidy level (defined as higher than tetraploidy; >4n), which is consistent with a frequency of 0.7–7.3% in previous publications (Ma et al., 2005; Troxell et al., 2006; Wang et al., 2002). Increasing CEN‐17 levels were statistically significantly associated with increasing risk of DFS events and mortality. When adjusting for patient and tumor characteristics including HER2 and TOP2A in a multivariate Cox regression analysis, CEN‐17 ploidy level was not an independent predictor of DFS or OS. We did not find any evidence for heterogeneity of the treatment effect according to ploidy level, e.g. CEF did not confer incremental benefit compared to CMF in patients with tumors of high ploidy level. Interestingly, we found that CMF confer a borderline significant incremental benefit compared to CEF in the patients with high CEN‐17 level but HER2 and TOP2A normal tumors, while a borderline significant improvement in OS was found among patients with HER2, TOP2A and CEN‐17 normal tumors if treated with CEF compared CMF. Thus, copy number changes of centromere 17 could not be established as an independent prognostic marker and were rarely observed in HER2 and TOP2A normal breast cancers. Increasing ploidy was, however, related to increased HER2 amplification and TOP2A deletion.
This study has some potential limitations. First, our study was not powered to examine a possible prognostic implication of very high ploidy levels and only 30 (3.9%) had tumors with a ploidy level higher than tetraploid (>4n). Second, dividing cancer cells may be misclassified since CEN‐17 FISH is unable to distinguish between aneuploidy and polyploidy in cells with three or four copies of CEN‐17. Third, it has not been clarified whether additional reference probes should be used to correct for dividing cells, e.g. CEN‐2 or CEN‐10 as chromosome 2 and 10 are the least affected in breast cancer (Mitelman, 2008) in contrast to CEN‐17 (Marchio et al., 2009; Troxell et al., 2006; Yeh et al., 2009).
The strengths of our study include the use of breast cancer tissue from participants in a large randomized trial and use of normal breast tissue to establish ploidy levels of cut sections, showing that that both the lack of whole nuclei in cut tissue sections and the localization of the target DNA sequence within the nucleus influence the “diploid” signal number. In contrast, earlier studies have used arbitrary cut‐offs (Dal Lago et al., 2006; Ma et al., 2005) or defined a normal range as the mean plus three standard deviations (Beser et al., 2007; Watters et al., 2003). Our results on the number of centromere and gene signals on normal breast tissue are very close to those reported on HER2 FISH negative samples with a HercepTest score of 0 (Lal et al., 2003).
Previously, the predictive value of TOP2A aberrations for benefit from anthracycline treatment has been shown (Knoop et al., 2005; Nielsen et al., 2008; O'Malley et al., 2009), while one study group (Bartlett et al., 2008b; Bartlett et al., 2010) found predictive value of the reference probe (centromere 17) rather than the gene probe (TOP2A). An association between an increase in centromere 17 and incremental benefit from anthracycline‐based chemotherapy was found both in the analysis of BR9601 alone (Bartlett et al., 2008b) and in a combined analysis of the NEAT and BR9601 trials (Bartlett et al., 2010) while HER2 and TOP2A status were not. However, there are factors in the NEAT/BR9601 study that makes direct comparison to other studies difficult. First, despite defining TOP2A amplification by a gene to centromere ratio of 1.5 or higher, the frequency of amplifications was lower than the frequency of deletions despite using a standard cut‐off for deletions. Secondly, HER2 amplification and TOP2A deletion were furthermore in univariate analysis of the NEAT/BR9601 study the only significant prognostic factors as opposed to the multivariate analysis where only CEN‐17 was an independent prognostic factor. Third, the multivariate analysis of NEAT/BR9601 is based on TOP2A normal compared to TOP2A abnormal (amplification and deletion combined) while the univariate analysis did not group TOP2A amplifications and deletions. We cannot confirm that incremental benefit of anthracycline‐based treatment is associated with centromere 17 copy number. The gene product of TOP2A is the direct target of anthracyclines (Gudkov et al., 1993) while the biological explanation for a specific anthracycline benefit in CEN‐17 duplicated tumors is lacking. CEN‐17 copy number might be a surrogate marker for TOP2A deletion and HER2 amplifications. Clarification of the complex rearrangements on chromosome 17 is expected from new techniques, i.e. zoom‐in aCGH has shown that amplification of the centromeric region of chromosome 17 frequently include the adjacent gene WSB1 on 17q11.2 (Staaf et al., 2010). WSB1 may be involved in the metastatic process (Archange et al., 2008), and it may be the driver gene rather that the centromeric non‐coding α‐satellite repeat. Probes that target the highly repetitive α‐satellite region of centromeres are much less prone to methodological variation and will often led to a signal even under suboptimal hybridization conditions. Alternatively, high ploidy levels may reflect insufficient DNA repair and could potentially lead to increased responsiveness to anthracyclines by a TOP2A independent mechanism (Burrell et al., 2010).
In the present study the ploidy levels were defined based on measurements in 120 normal breast samples The truncated nuclei of cut sections have fewer signals than whole nuclei (1.81 versus the expected 2.00 TOP2A signals) and more CEN‐17 than TOP2A signals are lost in the truncated nuclei (1.68 CEN‐17 signals versus 1.81 TOP2A signals) and thus leading to a TOP2A/CEN‐17 ratio of 1.08, slightly above the expected 1.00 in whole nuclei. The explanation for this phenomenon is that the non‐coding centromeres are preferentially observed near the membrane of the nucleus (Bolzer et al., 2005) while the coding gene signals are located more centrally in the nucleus. When preparing cut sections more centromere than gene signals are lost in the largest nuclei and fragments of nuclei that only contain a single centromere signal will often be excluded from the scoring.
The increasing diagnostic use of fluorescence in situ hybridization (FISH) for detection HER2 gene amplification in breast cancer patients (Wolff et al., 2007) has induced debate about the biological process behind the observed red and green signals in the truncated nuclei of the tissue sections available for diagnosis (reviewed by Reinholz et al., 2009). Extra copies of the green reference probe for centromere 17 have been taken as an indication of extra copies of whole chromosome 17 also referred to as polysomy 17. The definition of polysomy 17 is not uniform across different studies thus adding to confusion about the frequency and importance of polysomy 17, but the most widely accepted definition is three or more copies of CEN‐17 (Corzo et al., 2007; Reinholz et al., 2009; Wolff et al., 2007) although this cutoff is used both as an mean value and a count in at least 30% of the analyzed cells (Reinholz et al., 2009). Our study where standard cytogenetic definition of ploidy levels is transformed from whole nuclei to the truncated nuclei of the cut sections of paraffin‐embedded tissue supports the definition used in the HER2 guidelines (Wolff et al., 2007) using an mean of three or more CEN‐17 copies. Wolff et al. (2007) cite that polysomy 17 is observed in approximately 8% of all specimens and in line with other studies (Hyun et al., 2008; Ma et al., 2005; Merola et al., 2006; Reinholz et al., 2009; Wang et al., 2002) our data shows that high copy numbers of centromere 17 is relatively rare in breast tumors as only 9% had more than three centromere 17 signals.
Increased number of centromere 17 is observed in 8–68% of all tumor specimens (Reinholz et al., 2009), mostly among tumors with 4–6 HER2 gene copies (Bose et al., 2001; Downs‐Kelly et al., 2005; Ma et al., 2005; Vanden Bempt et al., 2008; Wolff et al., 2007). The large variation reflects the differences in definitions of polysomy/aneusomy 17 have been used. The highest frequency of polysomic tumors is reported in studies including elevated proportion of HER2 positive samples (Reinholz et al., 2009; Vanden Bempt et al., 2008; Watters et al., 2003). Less than 1% of tumor samples referred to HER2 FISH analysis (Press, 2006) has been reported polysomic, but this low frequency is obtained using a cutoff of >5 CEN‐17 per cell. Several studies further divided polysomy 17 into low‐level and high‐level polysomy and observed that low‐level polysomy 17 was the more common of the two (26–43% vs. 5–7%) (Bose et al., 2001; Hyun et al., 2008; Ma et al., 2005; Merola et al., 2006; Wang et al., 2002; Watters et al., 2003). Dividing cells are expected to contribute to the increased ploidy levels of breast tumors, but we did not find any correlation between ploidy level and tumor grade. High grade has been reported to correlate with polysomy 17 (Hyun et al., 2008; Nakopoulou et al., 2002; Watters et al., 2003), but it is not a general finding (Lal et al., 2003; Vanden Bempt et al., 2008). Three recent studies (Marchio et al., 2009; Moelans et al., 2010; Yeh et al., 2009) have demonstrated that whole chromosome 17 polysomy is rare and that those cases allegedly identified as chromosomes 17 polysomic by FISH may more correctly be termed “chromosome 17 centromere amplified”. Centromere 17 amplification has previously been reported by FISH for tumors with very high centromere 17 copy number (Troxell et al., 2006).
Increased centromere 17 copy numbers are not associated with elevated HER2 protein or mRNA levels (Corzo et al., 2007; Dal Lago et al., 2006; Downs‐Kelly et al., 2005; Lal et al., 2003; Ma et al., 2005; Vanden Bempt et al., 2008; Wang et al., 2002) although the HER2 gene numbers were elevated. These studies demonstrated the importance of measuring the ratio of gene to reference signals to clearly discriminate between true HER2 amplification and polyploidy as polysomic tumors without HER2 amplification resembles HER2‐negative tumors (Dal Lago et al., 2006; Vanden Bempt et al., 2008). Inclusion of centromere 17 reference probe has been reported to avoid an incorrect diagnosis of amplification in 2.81% of the patients tested (Bartlett et al., 2008a) while less than 1% is misclassified due to CEN‐17 amplification (Varga et al., 2011). Neither the degree of HER2 amplification nor polysomy 17 influences prognosis or benefits from HER2 targeting treatment (Downey et al., 2010; Dowsett et al., 2009; Perez et al., 2010; Press et al., 2008).
In conclusion, centromere 17 counts in normal breast tissue were used to define the ploidy level in samples from breast cancer patients as obtained from HER2 and TOP2A testing of patients from a large clinical trial. Amplification of HER2 and deletion of TOP2A were correlated with increasing ploidy. High centromere 17 level is not independently prognostic for disease‐free survival or overall survival and does not independently from TOP2A identify patients who are most likely to benefit from anthracyclines.
Conflict of interest
S Møller, M‐B Jensen, A Knoop, E Balslev and HT Mouridsen have no conflict of interest to declare. K Vang Nielsen and S Müller are employees of Dako A/S. B Ejlertsen has a relative who is employed by Dako A/S.
Acknowledgment
We thank Signe Lykke Nielsen for excellent technical assistance.
Vang Nielsen Kirsten, Ejlertsen Bent, Møller Susanne, Jensen Maj-Britt, Balslev Eva, Müller Sven, Knoop Ann, Mouridsen Henning T., (2012), Lack of independent prognostic and predictive value of centromere 17 copy number changes in breast cancer patients with known HER2 and TOP2A status, Molecular Oncology, 6, doi: 10.1016/j.molonc.2011.11.006.
Supported by grants from the Danish Ministry of Health (J No. 2006‐12103‐272) and from the Danish Research Council (J. No. 271060542).
References
- Archange, C. , Nowak, J. , Garcia, S. , Moutardier, V. , Calvo, E.L. , Dagorn, J.C. , Iovanna, J.L. , 2008. The WSB1 gene is involved in pancreatic cancer progression. PLoS ONE. 3, e2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, J.M. , Campbell, F.M. , Mallon, E.A. , 2008. Determination of HER2 amplification by in situ hybridization: when should chromosome 17 also be determined?. Am. J. Clin. Pathol.. 130, 920–926. [DOI] [PubMed] [Google Scholar]
- Bartlett, J.M. , Munro, A. , Cameron, D.A. , Thomas, J. , Prescott, R. , Twelves, C.J. , 2008. Type 1 receptor tyrosine kinase profiles identify patients with enhanced benefit from anthracyclines in the BR9601 adjuvant breast cancer chemotherapy trial. J. Clin. Oncol.. 26, 5027–5035. [DOI] [PubMed] [Google Scholar]
- Bartlett, J.M. , Munro, A.F. , Dunn, J.A. , McConkey, C. , Jordan, S. , Twelves, C.J. , Cameron, D.A. , Thomas, J. , Campbell, F.M. , Rea, D.W. , Provenzano, E. , Caldas, C. , Pharoah, P. , Hiller, L. , Earl, H. , Poole, C.J. , 2010. Predictive markers of anthracycline benefit: a prospectively planned analysis of the UK National Epirubicin Adjuvant Trial (NEAT/BR9601). Lancet Oncol.. 11, 266–274. [DOI] [PubMed] [Google Scholar]
- Beser, A.R. , Tuzlali, S. , Guzey, D. , Dolek Guler, S. , Hacihanefioglu, S. , Dalay, N. , 2007. HER-2, TOP2A and chromosome 17 alterations in breast cancer. Pathol. Oncol. Res.. 13, 180–185. [DOI] [PubMed] [Google Scholar]
- Bolzer, A. , Kreth, G. , Solovei, I. , Koehler, D. , Saracoglu, K. , Fauth, C. , Muller, S. , Eils, R. , Cremer, C. , Speicher, M.R. , Cremer, T. , 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol.. 3, e157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, S. , Mohammed, M. , Shintaku, P. , Rao, P.N. , 2001. Her-2/neu gene amplification in low to moderately expressing breast cancers: possible role of chromosome 17/Her-2/neu polysomy. Breast J.. 7, 337–344. [DOI] [PubMed] [Google Scholar]
- Burrell, R.A. , Juul, N. , Johnston, S.R. , Reis-Filho, J.S. , Szallasi, Z. , Swanton, C. , 2010. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J. Cell. Biochem.. 111, 782–790. [DOI] [PubMed] [Google Scholar]
- Corzo, C. , Bellosillo, B. , Corominas, J.M. , Salido, M. , Coll, M.D. , Serrano, S. , Albanell, J. , Sole, F. , Tusquets, I. , 2007. Does polysomy of chromosome 17 have a role in ERBB2 and topoisomerase IIalpha expression? Gene, mRNA and protein expression: a comprehensive analysis. Tumor Biol.. 28, 221–228. [DOI] [PubMed] [Google Scholar]
- Dal Lago, L. , Durbecq, V. , Desmedt, C. , Salgado, R. , Verjat, T. , Lespagnard, L. , Ma, Y. , Veys, I. , Di Leo, A. , Sotiriou, C. , Piccart, M. , Larsimont, D. , 2006. Correction for chromosome-17 is critical for the determination of true Her-2/neu gene amplification status in breast cancer. Mol. Cancer Ther.. 5, 2572–2579. [DOI] [PubMed] [Google Scholar]
- Dhesy-Thind, B. , Pritchard, K.I. , Messersmith, H. , O'Malley, F. , Elavathil, L. , Trudeau, M. , 2008. HER2/neu in systemic therapy for women with breast cancer: a systematic review. Breast Cancer Res. Treat.. 109, 209–229. [DOI] [PubMed] [Google Scholar]
- Downey, L. , Livingston, R.B. , Koehler, M. , Arbushites, M. , Williams, L. , Santiago, A. , Guzman, R. , Villalobos, I. , Di Leo, A. , Press, M.F. , 2010. Chromosome 17 polysomy without human epidermal growth factor receptor 2 amplification does not predict response to lapatinib plus paclitaxel compared with paclitaxel in metastatic breast cancer. Clin. Cancer Res.. 16, 1281–1288. [DOI] [PubMed] [Google Scholar]
- Downs-Kelly, E. , Yoder, B.J. , Stoler, M. , Tubbs, R.R. , Skacel, M. , Grogan, T. , Roche, P. , Hicks, D.G. , 2005. The influence of polysomy 17 on HER2 gene and protein expression in adenocarcinoma of the breast: a fluorescent in situ hybridization, immunohistochemical, and isotopic mRNA in situ hybridization study. Am. J. Surg. Pathol.. 29, 1221–1227. [DOI] [PubMed] [Google Scholar]
- Dowsett, M. , Procter, M. , McCaskill-Stevens, W. , de Azambuja, E. , Dafni, U. , Rueschoff, J. , Jordan, B. , Dolci, S. , Abramovitz, M. , Stoss, O. , Viale, G. , Gelber, R.D. , Piccart-Gebhart, M. , Leyland-Jones, B. , 2009. Disease-free survival according to degree of HER2 amplification for patients treated with adjuvant chemotherapy with or without 1 year of trastuzumab: the HERA Trial. J. Clin. Oncol.. 27, 2962–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejlertsen, B. , Mouridsen, H.T. , Jensen, M.B. , Andersen, J. , Cold, S. , Edlund, P. , Ewertz, M. , Jensen, B.B. , Kamby, C. , Nordenskjold, B. , Bergh, J. , 2007. Improved outcome from substituting methotrexate with epirubicin: results from a randomised comparison of CMF versus CEF in patients with primary breast cancer. Eur. J. Cancer. 43, 877–884. [DOI] [PubMed] [Google Scholar]
- Gennari, A. , Sormani, M.P. , Pronzato, P. , Puntoni, M. , Colozza, M. , Pfeffer, U. , Bruzzi, P. , 2008. HER2 status and efficacy of adjuvant anthracyclines in early breast cancer: a pooled analysis of randomized trials. J. Natl. Cancer Inst.. 100, 14–20. [DOI] [PubMed] [Google Scholar]
- Guano, F. , Pourquier, P. , Tinelli, S. , Binaschi, M. , Bigioni, M. , Animati, F. , Manzini, S. , Zunino, F. , Kohlhagen, G. , Pommier, Y. , Capranico, G. , 1999. Topoisomerase poisoning activity of novel disaccharide anthracyclines. Mol. Pharmacol.. 56, 77–84. [DOI] [PubMed] [Google Scholar]
- Gudkov, A.V. , Zelnick, C.R. , Kazarov, A.R. , Thimmapaya, R. , Suttle, D.P. , Beck, W.T. , Roninson, I.B. , 1993. Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proc. Natl. Acad. Sci. U. S. A.. 90, 3231–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, C.L. , Lee, H.E. , Kim, K.S. , Kim, S.W. , Kim, J.H. , Choe, G. , Park, S.Y. , 2008. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J. Clin. Pathol.. 61, 317–321. [DOI] [PubMed] [Google Scholar]
- ISCN, 2009. An International System for Human Cytogenetic Nomenclature Karger; Basel: [Google Scholar]
- Knoop, A.S. , Knudsen, H. , Balslev, E. , Rasmussen, B.B. , Overgaard, J. , Nielsen, K.V. , Schonau, A. , Gunnarsdottir, K. , Olsen, K.E. , Mouridsen, H. , Ejlertsen, B. , 2005. Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J. Clin. Oncol.. 23, 7483–7490. [DOI] [PubMed] [Google Scholar]
- Lal, P. , Salazar, P.A. , Ladanyi, M. , Chen, B. , 2003. Impact of polysomy 17 on HER-2/neu immunohistochemistry in breast carcinomas without HER-2/neu gene amplification. J. Mol. Diagn.. 5, 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo, A.D. , Desmedt, C. , Bartlett, J.M. , Piette, F. , Ejlertsen, B. , Pritchard, K.I. , Larsimont, D. , Poole, C. , Isola, J. , Earl, H. , Mouridsen, H. , O'Malley, F.P. , Cardoso, F. , Tanner, M. , Munro, A. , Twelves, C.J. , Sotiriou, C. , Shepherd, L. , Cameron, D. , Piccart, M.J. , Buyse, M. , 2011. HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol.. 12, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Lespagnard, L. , Durbecq, V. , Paesmans, M. , Desmedt, C. , Gomez-Galdon, M. , Veys, I. , Cardoso, F. , Sotiriou, C. , Di Leo, A. , Piccart, M.J. , Larsimont, D. , 2005. Polysomy 17 in HER-2/neu status elaboration in breast cancer: effect on daily practice. Clin. Cancer Res.. 11, 4393–4399. [DOI] [PubMed] [Google Scholar]
- Marchio, C. , Lambros, M.B. , Gugliotta, P. , Di Cantogno, L.V. , Botta, C. , Pasini, B. , Tan, D.S. , Mackay, A. , Fenwick, K. , Tamber, N. , Bussolati, G. , Ashworth, A. , Reis-Filho, J.S. , Sapino, A. , 2009. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J. Pathol.. 219, 16–24. [DOI] [PubMed] [Google Scholar]
- McShane, L.M. , Altman, D.G. , Sauerbrei, W. , Taube, S.E. , Gion, M. , Clark, G.M. , 2005. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst.. 97, 1180–1184. [DOI] [PubMed] [Google Scholar]
- Merola, R. , Mottolese, M. , Orlandi, G. , Vico, E. , Cognetti, F. , Sperduti, I. , Fabi, A. , Vitelli, G. , Cianciulli, A.M. , 2006. Analysis of aneusomy level and HER-2 gene copy number and their effect on amplification rate in breast cancer specimens read as 2+ in immunohistochemical analysis. Eur. J. Cancer. 42, 1501–1506. [DOI] [PubMed] [Google Scholar]
- Mitelman, F., 2008. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- Moelans, C.B. , de Weger, R.A. , van Diest, P.J. , 2010. Absence of chromosome 17 polysomy in breast cancer: analysis by CEP17 chromogenic in situ hybridization and multiplex ligation-dependent probe amplification. Breast Cancer Res. Treat.. 120, 1–7. [DOI] [PubMed] [Google Scholar]
- Nakopoulou, L. , Giannopoulou, I. , Trafalis, D. , Gakiopoulou, H. , Keramopoulos, A. , Davaris, P. , 2002. Evaluation of numeric alterations of chromosomes 1 and 17 by in situ hybridization in invasive breast carcinoma with clinicopathologic parameters. Appl. Immunohistochem. Mol. Morphol.. 10, 20–28. [DOI] [PubMed] [Google Scholar]
- Nielsen, K.V. , Ejlertsen, B. , Moller, S. , Jorgensen, J.T. , Knoop, A. , Knudsen, H. , Mouridsen, H.T. , 2008. The value of TOP2A gene copy number variation as a biomarker in breast cancer: update of DBCG trial 89D. Acta Oncol.. 47, 725–734. [DOI] [PubMed] [Google Scholar]
- Nielsen, K.V. , Müller, S. , Poulsen, T.S. , Gabs, S. , Schonau, A. , 2004. Combined use of PNA and DNA for fluorescence In Nielsen P.E., Situ Hybridization (FISH). second ed. Horizon Bioscience; Norfolk: 227–260. [Google Scholar]
- O'Malley, F.P. , Chia, S. , Tu, D. , Shepherd, L.E. , Levine, M.N. , Bramwell, V.H. , Andrulis, I.L. , Pritchard, K.I. , 2009. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. J. Natl. Cancer Inst.. 101, 615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.E. , Knudsen, H. , Rasmussen, B.B. , Balslev, E. , Knoop, A. , Ejlertsen, B. , Nielsen, K.V. , Schonau, A. , Overgaard, J. , 2004. Amplification of HER2 and TOP2A and deletion of TOP2A genes in breast cancer investigated by new FISH probes. Acta Oncol.. 43, 35–42. [DOI] [PubMed] [Google Scholar]
- Orsetti, B. , Nugoli, M. , Cervera, N. , Lasorsa, L. , Chuchana, P. , Ursule, L. , Nguyen, C. , Redon, R. , du Manoir, S. , Rodriguez, C. , Theillet, C. , 2004. Genomic and expression profiling of chromosome 17 in breast cancer reveals complex patterns of alterations and novel candidate genes. Cancer Res.. 64, 6453–6460. [DOI] [PubMed] [Google Scholar]
- Perez, E.A. , Reinholz, M.M. , Hillman, D.W. , Tenner, K.S. , Schroeder, M.J. , Davidson, N.E. , Martino, S. , Sledge, G.W. , Harris, L.N. , Gralow, J.R. , Dueck, A.C. , Ketterling, R.P. , Ingle, J.N. , Lingle, W.L. , Kaufman, P.A. , Visscher, D.W. , Jenkins, R.B. , 2010. HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. J. Clin. Oncol.. 28, 4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press, M.F. , 2006. How is Her-2/neu status established when Her-2/neu and chromosome 17 centromere are both amplified?. Am. J. Clin. Pathol.. 126, 673–674. [DOI] [PubMed] [Google Scholar]
- Press, M.F. , Finn, R.S. , Cameron, D. , Di Leo, A. , Geyer, C.E. , Villalobos, I.E. , Santiago, A. , Guzman, R. , Gasparyan, A. , Ma, Y. , Danenberg, K. , Martin, A.M. , Williams, L. , Oliva, C. , Stein, S. , Gagnon, R. , Arbushites, M. , Koehler, M.T. , 2008. HER-2 gene amplification, HER-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin. Cancer Res.. 14, 7861–7870. [DOI] [PubMed] [Google Scholar]
- Press, M.F. , Sauter, G. , Buyse, M. , Bernstein, L. , Guzman, R. , Santiago, A. , Villalobos, I.E. , Eiermann, W. , Pienkowski, T. , Martin, M. , Robert, N. , Crown, J. , Bee, V. , Taupin, H. , Flom, K.J. , Tabah-Fisch, I. , Pauletti, G. , Lindsay, M.A. , Riva, A. , Slamon, D.J. , 2011. Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J. Clin. Oncol.. 29, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholz, M.M. , Bruzek, A.K. , Visscher, D.W. , Lingle, W.L. , Schroeder, M.J. , Perez, E.A. , Jenkins, R.B. , 2009. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol.. 10, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon, D.J. , Clark, G.M. , Wong, S.G. , Levin, W.J. , Ullrich, A. , McGuire, W.L. , 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 235, 177–182. [DOI] [PubMed] [Google Scholar]
- Staaf, J. , Jonsson, G. , Ringner, M. , Vallon-Christersson, J. , Grabau, D. , Arason, A. , Gunnarsson, H. , Agnarsson, B.A. , Malmstrom, P.O. , Johannsson, O.T. , Loman, N. , Barkardottir, R.B. , Borg, A. , 2010. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res.. 12, R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon, A.K. , Clark, G.M. , Chamness, G.C. , Ullrich, A. , McGuire, W.L. , 1989. HER-2/neu oncogene protein and prognosis in breast cancer. J. Clin. Oncol.. 7, 1120–1128. [DOI] [PubMed] [Google Scholar]
- Troxell, M.L. , Bangs, C.D. , Lawce, H.J. , Galperin, I.B. , Baiyee, D. , West, R.B. , Olson, S.B. , Cherry, A.M. , 2006. Evaluation of Her-2/neu status in carcinomas with amplified chromosome 17 centromere locus. Am. J. Clin. Pathol.. 126, 709–716. [DOI] [PubMed] [Google Scholar]
- Vanden Bempt, I. , Van Loo, P. , Drijkoningen, M. , Neven, P. , Smeets, A. , Christiaens, M.R. , Paridaens, R. , De Wolf-Peeters, C. , 2008. Polysomy 17 in breast cancer: clinicopathologic significance and impact on HER-2 testing. J. Clin. Oncol.. 26, 4869–4874. [DOI] [PubMed] [Google Scholar]
- Varga, Z. , Tubbs, R.R. , Wang, Z. , Sun, Y. , Noske, A. , Kradolfer, D. , Bosshard, G. , Jochum, W. , Moch, H. , Ohlschlegel, C. , 2011. Co-amplification of the HER2 gene and chromosome 17 centromere: a potential diagnostic pitfall in HER2 testing in breast cancer. Breast Cancer Res. Treat.. [DOI] [PubMed] [Google Scholar]
- Viale, G. , 2009. Be precise! The need to consider the mechanisms for CEP17 copy number changes in breast cancer. J. Pathol.. 219, 1–2. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Hossein Saboorian, M. , Frenkel, E.P. , Haley, B.B. , Siddiqui, M.T. , Gokaslan, S. , Hynan, L. , Ashfaq, R. , 2002. Aneusomy 17 in breast cancer: its role in HER-2/neu protein expression and implication for clinical assessment of HER-2/neu status. Mod. Pathol.. 15, 137–145. [DOI] [PubMed] [Google Scholar]
- Watters, A.D. , Going, J.J. , Cooke, T.G. , Bartlett, J.M. , 2003. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res. Treat.. 77, 109–114. [DOI] [PubMed] [Google Scholar]
- Wolff, A.C. , Hammond, M.E. , Schwartz, J.N. , Hagerty, K.L. , Allred, D.C. , Cote, R.J. , Dowsett, M. , Fitzgibbons, P.L. , Hanna, W.M. , Langer, A. , McShane, L.M. , Paik, S. , Pegram, M.D. , Perez, E.A. , Press, M.F. , Rhodes, A. , Sturgeon, C. , Taube, S.E. , Tubbs, R. , Vance, G.H. , van de Vijver, M. , Wheeler, T.M. , Hayes, D.F. , 2007. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol.. 25, 118–145. [DOI] [PubMed] [Google Scholar]
- Wolman, S.R. , 1994. Fluorescence in situ hybridization: a new tool for the pathologist. Hum. Pathol.. 25, 586–590. [DOI] [PubMed] [Google Scholar]
- Yeh, I.T. , Martin, M.A. , Robetorye, R.S. , Bolla, A.R. , McCaskill, C. , Shah, R.K. , Gorre, M.E. , Mohammed, M.S. , Gunn, S.R. , 2009. Clinical validation of an array CGH test for HER2 status in breast cancer reveals that polysomy 17 is a rare event. Mod. Pathol.. 22, 1169–1175. [DOI] [PubMed] [Google Scholar]


