Figure 3.
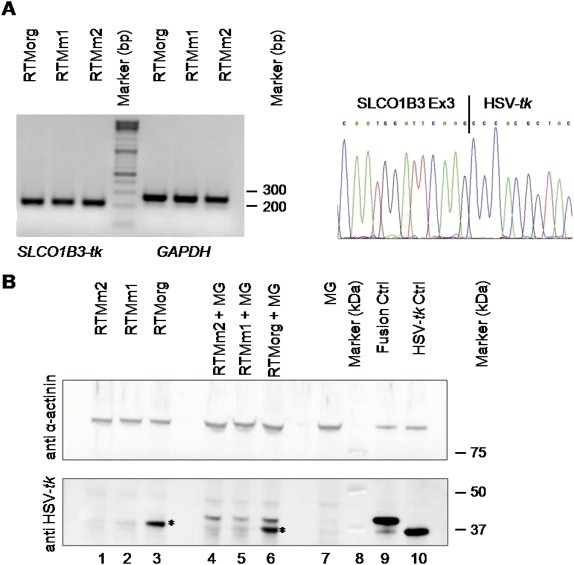
Detection of accurate trans‐splicing on the mRNA and protein level. (A) SqRT‐PCR analysis of HEK293AD cells double‐transfected with SLCO1B3‐MG and either the original RTM (RTMorg), or the mutated RTMs (RTMm1 and RTMm2). The correct PCR product of the fusion mRNA of SLCO1B3 and HSV‐tk (205 bp) is shown. GAPDH was used as housekeeping gene. All PCR products were verified by direct sequencing, demonstrating correct trans‐splicing between SLCO1B3‐MG and the RTMs on the mRNA level. A representative figure shows the sequence of the SLCO1B3‐HSV‐tk junction (right panel). (B) Sequence optimization leads to reduced background expression of RTMs as shown by Western blot analysis: Upper panel: α‐actinin staining (100 kDa) was used as a loading control. Lower panel: Western blot analysis demonstrated the expression of the SLCO1B3‐fusion protein (40 kDa) in all cell populations double‐transfected with the SLCO1B3‐MG (MG) and each RTM [lane 4, 5, 6]. Sequence optimized RTMm1 and RTMm2 showed greatly reduced background expression of HSV‐tk (37 kDa) in both single [lane 1, 2] and double [lane 4, 5] transfected HEK293 cells as compared to the RTMorg [lane 3, 6] (marked by asterisk). Cells transfected with the MG alone [lane 7], either the fusion [lane 9] or HSV‐tk [lane 10] construct served as negative and positive controls, respectively. Lane 8: Precision Plus Protein WesternC Standards (Biorad).
