Figure 1.
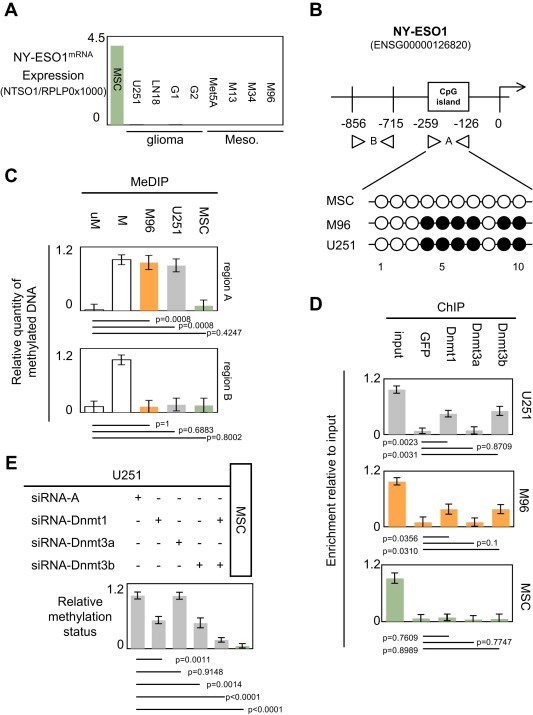
Dnmt1 and Dnmt3b participate to the epigenetic regulation of the NY‐ESO1 gene. (A) Expression level of NY‐ESO1mRNA in a panel of glioma and mesothelioma cells. Mesenchymal stem cell (MSC) was used as positive control i.e. cells expressing NY‐ESO1mRNA (Cronwright et al., 2005). U251 and LN18 are glioma cell lines, G1 and G2 are primary‐cultured tumor cells obtained from GBM, Met5A is mesothelial cell line, and M13, M34 and M96 are primary‐cultured tumor cells obtained from mesothelioma. (B) Schematic representation of the NY‐ESO1 gene. Methylated DNA immunoprecipitation experiments (MeDIP) were designed to estimate the methylation level of the −856/−715 and −259/−126 regions of the NY‐ESO1 gene i.e. one region including into CpG island and other not included into CpG island. Presence of CpG island was determined by using MethPrimer program. Methylation level of CG dinucleotides was determined by bisulfite sequencing (open circle: unmethylated, black circle: methylated). (C) Methylation status of the −856/−715 (bottom) and −259/−126 (top) regions of the NY‐ESO1 gene. uM: Unmethylated DNA: CpGenome™ Universal unmethylated DNA set, M: Methylated DNA : CpGenome™ Universal methylated DNA set, (Chemicon‐Millipore, France). (D) Identification by chromatin immunoprecipitation (ChIP) of the Dnmt recruited on the −259/−126 regions of the NY‐ESO1 gene. (E) Impact of the siRNA‐induced down‐regulation of Dnmt1, Dnmt3a and/or Dnmt3b on the methylation status of the −259/−126 region of the NY‐ESO1 gene. siRNA‐A is an aspecific siRNA used in control according to the manufacturer's instruction (Tebu‐Bio, Santa Cruz, France). The use of these siRNA was already monitored and described in previous report (Hervouet et al., 2010a).
