Figure 8. SCFA restore microglia malformation and immaturity in GF mice.
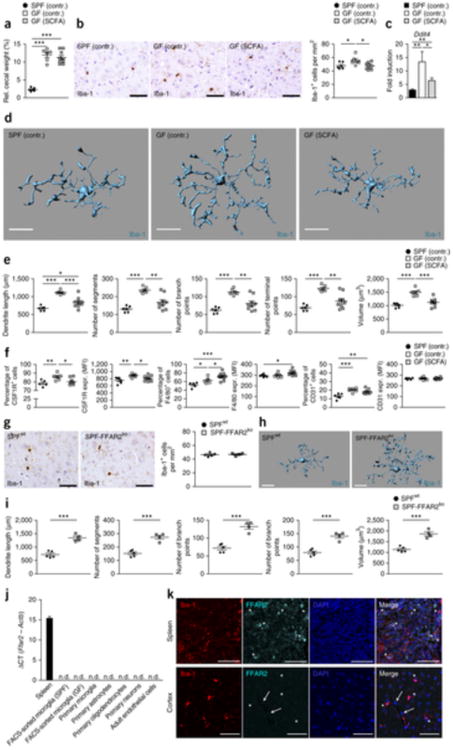
(a) Relative cecal weight of six SPF (SPF contr.), six GF mice fed with sodium-matched water (GF contr.) or nine GF mice treated with SCFAs (GF SCFA) for 4 weeks. Symbols represent data from individual mice. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (***P < 0.001). P values: SPF (contr.) versus GF (contr.), 0.0001; SPF (contr.) versus GF (SCFA.), <0.0001. (b) CNS histology of the cerebral cortex that was subjected to immunohistochemistry for Iba-1 to detect microglia (left) and quantification (right). Scale bars represent 50 μm. Representative pictures of six examined control mice and nine SCFA-treated mice are displayed, respectively. Each symbol represents one mouse and three to four sections per mouse were examined. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05). P values: SPF (contr.) versus GF (contr.), 0.0456; GF (contr.) versus GF (SCFA.), 0.0118. (c) Expression of Ddit4 mRNA measured by qRT-PCR in microglia isolated from SPF (contr.) (black bar), GF (contr.) (white bar) or GF mice treated with SCFA (gray bar). Data are presented as mean ± s.e.m. with six control samples and nine SCFA-treated samples. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01). P values: SPF (contr.) versus GF (contr.), 0.0064; GF (contr.) versus GF (SCFA.), 0.0306; SPF (contr.) versus GF (SCFA.), 0.0056. (d) Morphology of representative cortical microglia (scale bars represent 15 μm, d) and Imaris-based quantification of cellular parameters (e). Each symbol displays one mouse with three measured cells per animal. Six SPF and GF control animals and nine SCFA-treated GF animals were investigated. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: dendrite length SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0008; SPF (contr.) versus GF (SCFA), 0.0247; number of segments SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0021; number of branch points SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0027; number of terminal points SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0018; volume SPF (contr.) versus GF (contr.), 0.0001; GF (contr.) versus GF (SCFA), 0.0004. (f) Quantifications of the percentages of positively labeled cells and MFIs for CSF1R, F4/80 and CD31 on microglia from six SPF (contr.), six GF (contr.) and nine GF (SCFA) mice. Each symbol represents one mouse. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (*P < 0.05, **P < 0.01, ***P < 0.001). P values: CSF1R (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0094; GF (contr.) versus GF (SCFA), 0.0156; CSF1R (MFI) SPF (contr.) versus GF (contr.), 0.0076; GF (contr.) versus GF (SCFA), 0.0427; F4/80 (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0204; GF (contr.) versus GF (SCFA), 0.0367; SPF (contr.) versus GF (SCFA), 0.0002; F4/80 (MFI) SPF (contr.) versus GF (SCFA), 0.0194; CD31 (percentage of positive cells) SPF (contr.) versus GF (contr.), 0.0009; SPF (contr.) versus GF (SCFA), 0.0023. (g) Iba-1 immunhistochemistry (left) and quantification (right) of cortical sections from four FFAR2-deficient (SPF-FFAR2ko) or five competent (SPFwt) mice. Scale bars represent 50 μm. Representative pictures are displayed. Each symbol represents one mouse. Three to four sections per mouse were examined. Data are presented as mean ± s.e.m. (h,i) Imaris-based three-dimensional reconstruction of representative microglia from SPF-FFAR2ko and (SPFwt) mice (scale bars represent 15 μm) and quantification of the cellular parameters (i). Each symbol shows one mouse with at least three measured cells per animal. Four FFAR2-deficient (SPF-FFAR2ko) and five competent (SPFwt) mice were analyzed. Data are presented as mean ± s.e.m. Significant differences were determined by an unpaired t test (***P < 0.001). P values: dendrite length, 0.0007; number of segments, 0.0007; number of branch points, 0.0006; number of terminal points, 0.0008; volume, 0.0008. (j) Ffar2 mRNAs levels measured by qRT-PCR for the indicated cells and tissues, respectively. Data are expressed as ΔCT ratio of Ffar2 mRNA expression versus endogenous Actb and exhibited as mean ± s.e.m. Bar represents means ± s.e.m. with at least three samples in each group. n.d., not detectable. (k) Immunofluorescence images of FFAR2 (aqua green) on Iba-1+ (red) microglia or macrophages of wild-type mice. Asterisks indicate double-positive cells in the spleen (upper row) and FFAR2-negative microglia in the cortex (lower row) of wild-type mice. Arrows highlight FFAR2-negative endothelial cells. Nuclei were stained with DAPI (blue). Scale bars represent 50 μm.
