Figure 1.
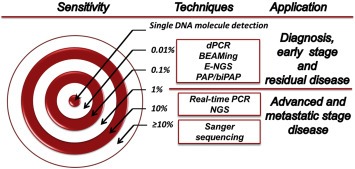
Different ctDNA detection techniques and their sensitivity. Techniques are ranked according to their usual sensitivity towards minor allele detection. The optimal technique would allow the detection of a single mutated DNA molecule whatever the normal DNA background. Digital PCR (dPCR), and its variants, droplet‐dPCR and BEAMing (“Beads, Emulsion, Amplification, Magnetic”), can be considered as the current standard for hotspot mutation detection (typically, KRAS mutations) in ctDNA, with 0.01%–0.1% sensitivity. Pyrophosphorolysis‐activated polymerization (PAP)‐PCR and bidirectional‐PAP (biPAP)‐PCR techniques have similar theoretical 0.01%–0.1% sensitivity (although not tested on KRAS). Modified next generation sequencing procedures (CAPP‐Seq, Safe‐Seq, etc, called “E‐NGS” here) exhibited at least 0.1% sensitivity.
