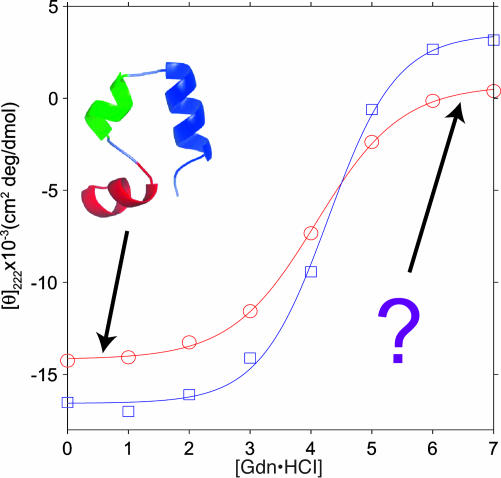
Fig. 1.
Chemical denaturation of HP35 by Gdn·HCl at 20°C (○) and -40°C (□), monitored by the CD signal at 222 nm. The known folded HP35 conformation at low [Gdn·HCl] is shown as a ribbon diagram. Solid-state NMR data shown in Figs. 2, 3, 4 probe the unknown conformational distributions at high [Gdn·HCl]. Solid lines are least-squares fits to a standard, simple, two-state model (40): CD(g) = CDfold - (CDfold - CDunf){exp[(mg - ΔG0)/RT]/{1 + exp[(mg - ΔG0)/RT]}}, where CDfold and CDunf are the CD signals of the folded and unfolded states, g · [Gdn·HCl], and ΔG0 is the free energy of unfolding at g = 0. At 20°C, m = 0.83 ± 0.10 kcal/mol·M (1 cal = 4.18 J), ΔG0 = 3.41 ± 0.40 kcal/mol, CDfold = -14,180 ± 320 cm2·deg/dmol, and CDunf = 710 ± 46 cm2·deg/dmol. At -40°C, m = 0.82 ± 0.21 kcal/mol·M, ΔG0 = 3.51 ± 0.90 kcal/mol, CDfold = -16,570 ± 810 cm2·deg/dmol, and CDunf = 3,510 ± 1,230 cm2·deg/dmol. At 4°C (data not shown), m = 0.79 ± 0.06 kcal/mol·M, and ΔG0 = 3.36 ± 0.25 kcal/mol. At -20°C (data not shown), m = 0.71 ± 0.07 kcal/mol·M, and ΔG0 = 3.17 ± 0.31 kcal/mol. (Errors are 95% confidence limits.)