ABSTRACT
Autophagy is an evolutionarily conserved catabolic process and is involved in the regulation of programmed cell death during the plant immune response. However, mechanisms regulating autophagy and cell death are incompletely understood. Here, we demonstrate that plant Bax inhibitor-1 (BI-1), a highly conserved cell death regulator, interacts with ATG6, a core autophagy-related protein. Silencing of BI-1 reduced the autophagic activity induced by both N gene-mediated resistance to Tobacco mosaic virus (TMV) and methyl viologen (MV), and enhanced N gene-mediated cell death. In contrast, overexpression of plant BI-1 increased autophagic activity and surprisingly caused autophagy-dependent cell death. These results suggest that plant BI-1 has both prosurvival and prodeath effects in different physiological contexts and both depend on autophagic activity.
KEYWORDS: ATG6, autophagy, Bax inhibitor-1, programmed cell death, Tobacco mosaic virus
Introduction
Autophagy is an evolutionarily conserved and tightly regulated catabolic process that maintains cell homeostasis by recycling unnecessary proteins and organelles to the lysosome or vacuole.1-3 Dysfunction of autophagy is associated with cancer, neurodegeneration, microbial infection, and aging.4 Previous studies in yeast and mammals have revealed most of the mechanisms underlying this process, central to which is the induction of autophagy, the formation of autophagosomes, their fusion with lysosomes or vacuoles.5 These sequential steps are achieved by autophagy-related (ATG) proteins.6
Plant ATG6 is the ortholog of yeast Vps30/Atg6, and mammalian BECN1/Beclin 1, which is also called Beclin 1 in tobacco plants. Mammalian BECN1 is a core ATG protein that contains a BCL2 (BCL2, apoptosis regulator)-homology-3 (BH3) domain, a central coiled-coil domain, and an evolutionarily conserved domain.7 It is part of the phosphatidylinositol 3-kinase (PtdIns3K) complex and positively regulates vesicle nucleation during autophagosome formation.8 In plants, only class III PtdIns3K is identified, which is encoded by a single copy gene, AT1G60490/PI3K/VPS34 in Arabidopsis,9 and AY701317/PI3K in tobacco.10 Recently, several BECN1-binding proteins have been characterized and an interactome has been shown to support BECN1s function during autophagy.11 BECN1-binding partners include the components of BECN1-PtdIns3K complexes, BCL2 family members, and other receptor and adaptor proteins, such as the ITPR (inositol 1,4,5-trisphosphate receptor).11 Very recently, β-tubulin was shown to interact with ATG6 in plants.12
BECN1 interacts with BCL2 family members to regulate autophagy in mammals.13 BCL2 family members, including BCL2, BCL2L1/ BCL-XL (BCL2 like 1), and viral BCL2 like proteins, suppress autophagy by inhibiting BECN1.7, 14-16 Phosphorylation of BCL2, BCL2L1 or BECN1 by MAPK8/JNK1 (mitogen-activated protein kinase 8), or DAPK (death associated protein kinase) disrupts the interaction, thus promoting autophagy.17,18 However, the antiapoptotic BCL2 family members were reported recently to affect autophagy and cell death indirectly, by inhibiting proapoptotic proteins BAX (BCL2 associated X) and BAK1 (BCL2 antagonist/killer 1).19 Recently a study in Arabidopsis suggests that the BCL2-associated athanogene family (BAG) member BAG6 functions in fungal resistance and autophagy in plants.20
Although plant autophagy has been reported to play important roles in nutrient recycling, biotic and abiotic stress responses, and development,21-28 little is known about the regulatory network underlying plant autophagy. Plant ATG6 is required for the induction of autophagy and autophagy regulates programmed cell death during the plant immune response.10,29-33 However, no BCL2 or BCL2L1 orthologs have been identified in plants, raising the question of how ATG6-dependent autophagy is regulated in plants.
Bax inhibitor-1 (BI-1), first identified as an inhibitor of BAX-induced cell death,34 is a highly evolutionarily conserved cell death regulator with multiple transmembrane domains. BI-1-like proteins are present in viruses, bacteria, yeast, plants, insects, and mammals.34-37 In mammals, TMBIM6/BI-1 (transmembrane BAX inhibitor motif containing 6) interacts with BCL2 and BCL2L1 to suppress mammalian apoptosis, probably by regulating endoplasmic reticulum (ER) calcium homeostasis.34,38-40 Besides inhibiting BAX-triggered cell death, mammalian TMBIM6 also inhibits ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1)-dependent unfolded protein response (UPR) by physically interacting with the ER stress sensor, ERN1, which is an ER resident serine/threonine-protein kinase/endoribonuclease, leading to increased susceptibility to mild ER stress.41 Thus, mammalian TMBIM6 may modulate cell death by affecting UPR or ER calcium homeostasis.42 In Arabidopsis, ER stress triggers autophagy and IRE1B is required for the ER stress-induced autophagy.24 A recent study suggests that accumulation of unfolded protein in ER will induce autophagy.43 BI-1 has also been characterized as a cell death suppressor and a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants.44-51 Arabidopsis BI-1 modulates ER stress-mediated programmed cell death without affecting the expression of UPR target genes.41,52 In addition, plant BI-1 regulates cell death by altering reactive oxygen species (ROS) level, Ca2+ flux control and lipid dynamics.53-55 In this study, we reveal that BI-1 functions not only as the well-recognized prosurvival factor, but also as a prodeath factor.
TMBIM6/BI-1 is described as a negative modulator of autophagy in cells undergoing nutrient starvation by controlling the ERN1-MAPK8 pathway through its inhibitory effect on ERN1.41,56 However, TMBIM6 is recently reported to regulate Ca2+-mediated bioenergetics to promote autophagy in an ITPR-dependent manner, leading to the controversial roles of BI-1 in autophagy.57 In this study, we show that plant BI-1 positively regulates autophagy by interacting with ATG6 and that autophagy is required for both the prosurvival and prodeath roles of BI-1.
Results
Identification and structural analysis of plant Bax inhibitor-1 as an ATG6-interacting partner
ATG6 regulates programmed cell death (PCD) during the plant innate immune response.10,29 To identify ATG6-binding proteins in plants, we performed a yeast 2-hybrid screen using Nicotiana tabacum ATG6 as bait and an Arabidopsis cDNA library as prey. In this screen, we identified Arabidopsis thaliana Bax inhibitor-1 (AtBI-1) as an interactor of ATG6. Next, we cloned N. benthamiana BI-1 (NbBI-1) cDNA and the encoded protein has a predicted length of 249 amino acids (aa), with high identity to orthologs from other organisms (43% to 97%). NbBI-1 also contains the conserved C-terminal 14 aa fragment (Fig. S1A) which is important for the interaction of BI-1 with other proteins in Arabidopsis and mammals.41,53-55
Mammalian ATG6 (BECN1) has a BH3 domain, which is defined as 4-turn amphipathic α-helices and contains the sequence feature as Hy-X-X-X-Hy-X-X-X-Sm-D/E-X-Hy, where Hy represents hydrophobic residues and Sm represents small residues.13 Similarly, tobacco (Nt) and Arabidopsis (At) ATG6s are also predicted to have a putative BH3 domain (Fig. S1B and C), and have an evolutionarily conserved domain (ECD) that is found in the crystallography of human ATG6 (PDB code: 4DDP) (Fig. S1D).58
Subcellular localization of BI-1 and ATG6 in plants
Mammalian and yeast orthologs of plant BI-1s (mammalian TMBIM6 and yeast BXI1 respectively) both exhibit an intramembrane distribution pattern, typically localizing to the ER and nuclear envelope.34,59 To determine the subcellular localization of tobacco BI-1 in plants, we generated a BI-1-YFP fusion construct driven by the CaMV 35S promoter. BI-1-YFP or YFP alone was transiently coexpressed with a CFP-tagged version of the ER marker, ER-cb, in N. benthamiana. ER-cb labels the cortical ER and perinuclear structure.60 BI-1-YFP mainly localized to the ER and perinuclear regions labeled by ER-cb (Fig. 1, first row). In addition, we found that ER-cb-labeled ER tubules were disrupted and formed clusters of large vesicle-like structures, after the expression of BI-1-YFP (Fig. 1, first row, as arrows indicate). BI-1-YFP localized to these punctate structures in these cells (Fig. 1, first row). As a control, YFP alone was freely distributed to the nucleus and cytoplasm and had no punctate structure on the ER (Fig. 1, third row).
Figure 1.
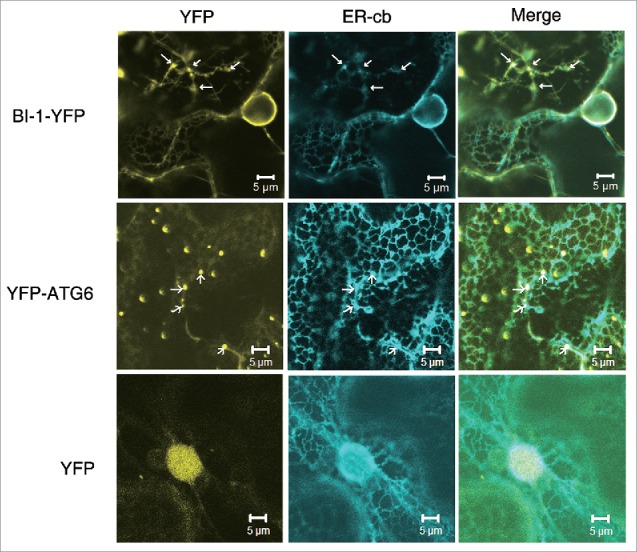
The subcellular localization of tobacco BI-1 and ATG6. BI-1-YFP localized to the ER network and nuclear envelope region in N. benthamiana cells (first row) and formed clusters of large vesicle-like punctate structures (arrows). YFP-ATG6 localized to the ER network, but strongly labeled punctate structures along the ER network (second row, arrows). The ER tubular network was labeled with the ER marker, ER-cb. YFP alone localized to the nucleus and cytoplasm and its expression had no effects on the ER morphology (third row), which was used as a control. Experiments were repeated 3 times.
Next, we tested whether the punctate structure of BI-1 was localized to the Golgi body. For this purpose, BI-1-YFP or YFP control was transiently coexpressed with the CFP-tagged Golgi marker, G-cb.60 BI-1-YFP mainly showed typical ER network localization and the punctate structures did not colocalize with Golgi bodies (Fig. S2A, first and second rows). These results suggest that plant BI-1 localizes to the ER and perinuclear regions.
Since BI-1 interacted with ATG6, we next examined the subcellular localization of ATG6. We generated YFP-ATG6 fusion construct driven by the CaMV 35S promoter, and then coexpressed this construct with ER-cb or G-cb. YFP-ATG6 weakly labeled the ER network, but strongly labeled punctate structures. In the cells expressing YFP-ATG6, most ER tubules maintained a network structure and some formed puncta labeled by ER-cb along the ER network, which merged with the YFP-ATG6 puncta (Fig. 1, second row). These puncta did not colocalize with the Golgi apparatus either (Fig. S2A, third row). When expressed alone, YFP-ATG6 formed punctate structures as well (Fig. S2B). Thus, tobacco ATG6 is partially distributed to the ER network, but mostly located in ER-associated puncta.
BI-1 interacts with ATG6 in vitro and in vivo
Since Arabidopsis BI-1 was found to interact with tobacco ATG6 in our yeast 2-hybrid screen, we tested whether tobacco BI-1 interacted with ATG6 in dual-membrane yeast 2-hybrid assay. Tobacco ATG6 was expressed in yeast as a fusion to the NubG domain (NubG-ATG6). BI-1 was expressed as a fusion to Cub-LexA-VP16, (BI-1-Cub-LexA-VP16). Upon interaction, LexA-VP16 is cleaved and translocated to the nucleus to activate 3 reporter genes, i.e., HIS3, ADE2, and lacZ. NubG-ATG6-containing yeast transformed with BI-1-Cub-LexA-VP16, but not with the empty vector (EV), turned blue on plates containing 5-bromo-4-chloro-3-indolyl-D-galactoside (X-gal) and grew on plates lacking His and Ade (Fig. 2A), suggesting that tobacco BI-1 interacts with tobacco ATG6 in yeast.
Figure 2.
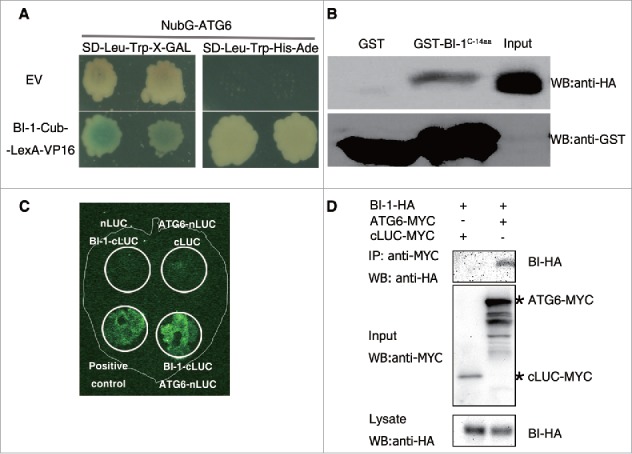
Tobacco BI-1 interacted with ATG6 in vitro and in vivo. (A) A split ubiquitin yeast 2-hybrid assay showed the interaction between BI-1 and ATG6. NubG-ATG6-harboring yeast strains transformed with BI-1-Cub-LexA-VP16, but not Cub-LexA-VP16, turned blue on X-gal plates (SD-Leu-Trp-X-Gal) and grew on medium lacking His and Ade (auxotrophic assay, SD-Leu-Trp-His-Ade). (B) GST affinity isolation assay showed the interaction between BI-1 and ATG6. A GST fusion with the C-terminal 14 aa of BI-1 (GST-BI-1C-14aa) or GST immobilized on glutathione-sepharose beads were incubated with the HA-tagged N-terminal 1–192 aa domain of ATG6 (HA-ATG61–192), which was generated in recombinant E. coli. Beads were washed and proteins were separated by SDS-PAGE. Immunoblot assays were performed with an anti-HA or anti-GST antibody. (C) LCI assay showed the interaction between BI-1 and ATG6 in vivo. Pairs of proteins were coexpressed in different areas (circles) of the same N. benthamiana leaf. Combinations of nLUC+BI-1-cLUC and ATG6-nLUC+cLUC were used as negative controls, while Arabidopsis SGT1A-nLUC+cLUC-RAR1 was used as the positive control. (D) Coimmunoprecipitation of BI-1-HA and ATG6-MYC. BI-1-HA with ATG6-MYC or cLUC-MYC (control) were coexpressed in N. benthamiana respectively and immunoprecipitated with anti-MYC beads. Extracts and precipitates were analyzed by western blot using indicated antibodies.
We sought to confirm the interaction of BI-1 with ATG6 using a glutathione S-transferase (GST) affinity isolation assay. However, we were unable to express the soluble full-length BI-1 and ATG6 proteins in E. coli. Since the C-terminal 14 aa of Arabidopsis BI-1 is the putative cytoplasmic part of BI-1 and the N-terminal domain of ATG6, which contains a predicted BH3 domain (Fig. S1B and C), is required for its interaction with BCL2 family members,7,16,55,61 we tested whether there was a direct interaction between these regions. We expressed GST-tagged C-terminal 14 aa of BI-1 (GST-BI-1C-14aa), GST alone and an HA-tagged fusion of the N-terminal 192 aa of tobacco ATG6 (HA-ATG61–192) in E. coli. The GST affinity isolation assay showed that GST-BI-1C-14aa, but not GST alone, bound directly to HA-ATG61–192 (Fig. 2B), suggesting that the C-terminal 14 aa of tobacco BI-1 interact directly with the N-terminal 192 aa of tobacco ATG6.
Furthermore, we performed a firefly luciferase complementation imaging (LCI) assay to test the interaction of BI-1 with ATG6 in plants.62 Tobacco BI-1 was fused to the C-terminal domain of luciferase (cLUC), while tobacco ATG6 was fused to the N-terminal domain of luciferase (nLUC). ATG6-nLUC was coexpressed with BI-1-cLUC in N. benthamiana. Upon interaction, luciferase is reconstituted and can be readily detected as a chemical luminescent signal.62 Signals were detected when ATG6 was coexpressed with BI-1 (Fig. 2C).
We also performed coimmunoprecipitation assays to test the interaction in plants. Tobacco BI-1 with HA tag (BI-1-HA), coimmunoprecipitated with cMYC tagged tobacco ATG6 (ATG6-MYC) but not the negative control cLUC-MYC (Fig. 2D).
Taken together, these results, with bimolecular fluorescence complementation (BiFC) below, suggest that plant BI-1 interacts with ATG6 and that the C-terminal 14 aa of BI-1 is sufficient for the interaction.
BI-1 and ATG6 interacted at the ER
The subcellular localization of the BI-1-ATG6 interaction in plants was investigated using a Citrine YFP-based BiFC assay.63 Tobacco BI-1 was fused to the N-terminal domain of YFP (nYFP) to generate BI-1-nYFP, and tobacco ATG6 was fused to the C-terminal domain of YFP (cYFP) to generate cYFP-ATG6. cYFP-ATG6 was transiently coexpressed in N. benthamiana with BI-1-nYFP, along with the ER marker, ER-cb, or the Golgi marker, G-cb. A positive interaction signal indicated by yellow fluorescence was observed only when cYFP-ATG6 was combined with BI-1-nYFP (Fig. 3). Interestingly, the interaction between BI-1-nYFP and cYFP-ATG6 yielded fluorescent punctate structures that merged with the ER-cb signals, indicating that these proteins interacted at the ER (Fig. 3, first row and second row). The negative control, BI-1-nYFP plus cYFP-nLUC, showed no interaction signal (Fig. 3, third row and fourth row). In addition, the interaction signals of BI-1 and ATG6 did not colocalize with the G-cb-labeled Golgi (Fig. S3). These results suggest that plant BI-1 interacts with ATG6 at the ER.
Figure 3.
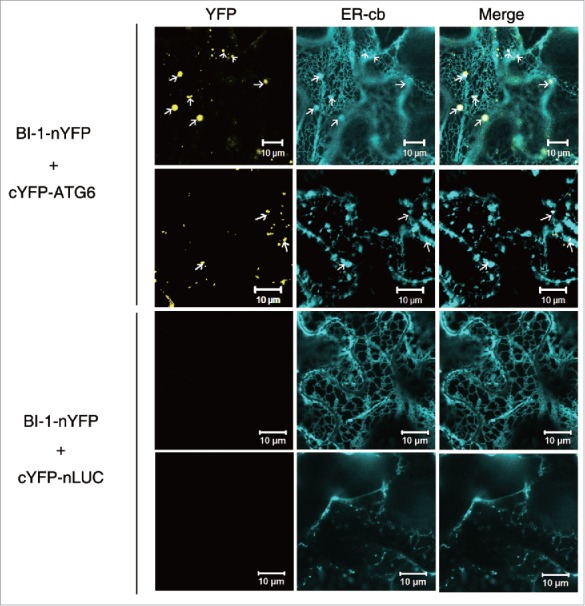
Tobacco BI-1 and ATG6 interacted at the ER. nYFP and cYFP are the N- and C-terminal domains of YFP, respectively. BI-1 was fused to the N-terminal end of nYFP. ATG6 was fused to the C-terminal end of cYFP. Yellow puncta in the first column indicate interactions between ATG6 and BI-1 and colocalize with puncta (first row and second row, arrows) labeled with the ER marker, ER-cb. No interaction was detected in the BI-1-nYFP+cYFP-nLUC negative control (third row and fourth row). Experiments were repeated 3 times.
Silencing of BI-1 compromised autophagy induced by N gene-mediated resistance to TMV
We used Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) to investigate the role of plant BI-1 in autophagy.64 Fragments of 2 NbBI-1 cDNAs (i.e., NbBI-1.1 and NbBI-1.2) were amplified by RT-PCR. The encoded proteins are 97% identical to each other (Fig. S4A). Because N. benthamiana is an allotetraploid, we reasoned that NbBI-1.1 and NbBI-1.2 were alleles of the same gene in a distant ancestor. N. benthamiana plants infected with a TRV vector carrying the NbBI-1.1 fragment (VIGS-BI-1) did not show any visible developmental phenotype compared with TRV alone nonsilenced control (Control). Real-time RT-PCR indicated that silencing of BI-1 reduced the mRNA level of BI-1 by 82%, but had no effect on mRNA level of BI-2, which encodes a protein sharing 84% protein identity with BI-1.1 (Fig. S4B), suggesting that silencing of BI-1 is effective and gene-specific.
E64d is a cysteine protease inhibitor that can increase the number of autophagic compartments by preventing their vacuole degradation. Thus, hereafter, all autophagy-related experiments were performed in the presence of E64d without specific statement. Monodansylcadaverine (MDC) is an autofluorescent substance that is commonly used to detect autophagic activity in plants and other organisms.65 We identified a few MDC-stained autophagic structures in nonsilenced control plants, suggesting that the basal autophagic activity is low in N. benthamiana plants. Furthermore, we found no obvious difference in the autophagic activity between the nonsilenced control and BI-1-silenced plants (Fig. S5A), which may be due to low level of basal autophagic activity.
We next evaluated the effect of silencing of BI-1 on the autophagic activity induced by the N gene-mediated resistance to TMV.10 As expected, we readily observed MDC-stained autophagic structures in the leaves of nonsilenced control N gene-containing N. benthamiana (NN) plants at 10 d postinfection (dpi) with GFP-tagged TMV (TMV-GFP). However, there were fewer MDC-stained autophagic structures in the BI-1-silenced NN plants than in the nonsilenced control NN plants (Fig. 4A and B).
Figure 4.
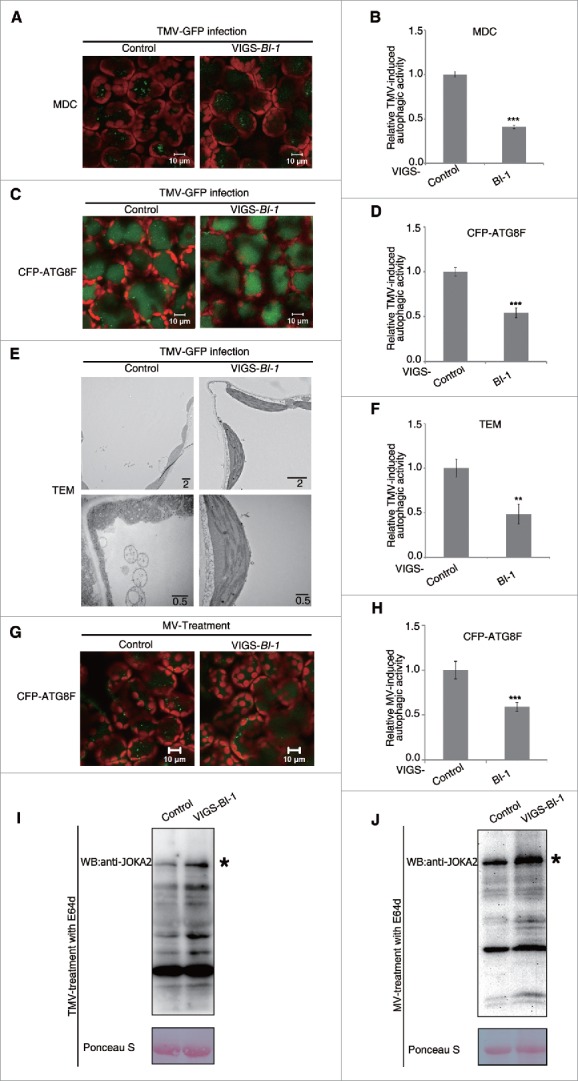
Silencing of BI-1 compromised autophagy induced by N gene-mediated resistance to TMV or MV. (A) Representative images of MDC-stained autophagic structures in TMV-infected leaf tissues of nonsilenced control (Control) and BI-1-silenced (VIGS-BI-1) N gene-containing plants at 10 dpi. The green fluorescent puncta were autophagic structures inside the vacuole and chloroplast autofluorescence was in red. Scale bars: 10 μm. (B) The relative TMV-induced autophagic activity in MDC-stained tissue. The number of MDC-stained autophagic structures per leaf section was counted and normalized to that of the Control. Data are the means±SEM of relative autophagic activity, with the mean value of the Control set to 1.0 (***P < 0.001, n = 10). (C) Representative images of CFP-ATG8F-labeled autophagic structures in TMV-infected leaf tissues of Control and VIGS-BI-1 N gene-containing plants at 8 dpi. The images were taken 2 d after agroinfiltration with CFP-ATG8F. The green fluorescent puncta were autophagic structures and chloroplast autofluorescence was in red. Scale bars: 10 μm. (D) The relative TMV-induced autophagic activity in CFP-ATG8F-labeled tissue. The number of CFP-ATG8F-labeled autophagic structures per leaf section was counted and normalized to that of Control. Data are the means ± SEM of the relative autophagic activity, with the mean value of the Control set to 1.0 (***P < 0.001, n = 10). (E) Representative transmission electron microscopy (TEM) images of TMV-infected leaf tissues from Control and VIGS-BI-1 N gene-containing plants at 10 dpi. Lower panel was TEM images of autophagosome shown at higher magnification. Scale bars are in μm. (F) The relative TMV-induced autophagic activity as determined in TEM assays. The number of autophagic structures per 100 μm2 was counted and normalized to that of Control. Data are the means ± SEM of relative autophagic activity, with the mean value of the Control set to 1.0 (**P < 0.01, n = 10). (G) Representative images of CFP-ATG8F-labeled autophagic structures in the MV-treated leaf tissues of Control and VIGS-BI-1 plants. The images were taken 2 d after agroinfiltration with CFP-ATG8F. The green fluorescent puncta were autophagic structures inside the vacuole and chloroplast autofluorescence was in red. (H) The relative MV-induced autophagic activity in CFP-ATG8F-labeled tissue. The number of CFP-ATG8F-labeled autophagic structures per leaf section was counted and normalized to that of Control. Data are the means±SEM of the relative autophagic activity, with the mean value of the Control set to 1.0 (***P < 0.001, n = 10). (I) BI-1-silenced plant accumulated more JOKA2 than control plant under the condition of TMV-induced autophagy. Ponceau S staining was used as loading control. (J) BI-1-silenced plants accumulated more JOKA2 than control plant under the condition of MV-induced autophagy. Ponceau S staining was used as loading control. Experiments were repeated at least 2 times. All autophagy-related experiments were performed with E64d treatment.
Fluorescent protein-tagged ATG8 is a useful marker to monitor macroautophagy in several organisms including plants.66 Transient expression of CFP-tagged ATG8F (CFP-ATG8F) in nonsilenced control and in BI-1-silenced NN plants revealed no difference in autophagy (Fig. S5B). We then transiently expressed CFP-ATG8F in TMV-infected NN leaf tissues and found that BI-1-silenced NN plants contained fewer CFP-ATG8F-labeled autophagic structures than the nonsilenced control plants in either the presence (Fig. 4C and D) or the absence of E64d (Fig. S6A and B).
We also used transmission electron microscopy (TEM) to verify the autophagy induced by N gene-mediated resistance to TMV. In TMV-infected nonsilenced control NN plants, we frequently observed autophagic structures containing electron-dense material in the cytoplasm and vacuole (Fig. 4E, control panel). However, we detected far fewer autophagic structures in BI-1-silenced NN plants (Fig. 4E and F).
NBR1 is an autophagy cargo receptor containing light chain 3 (LC3)- and ubiquitin (Ub)-binding domains in mammals.67 Plant NBR1, which is also called JOKA2 in tobacco,68 is identified as a selective autophagy cargo receptor recently, and itself is degraded in the vacuole once autophagy activity increases,69,70 enabling it as a useful tool to measure autophagy flux. Under the condition of TMV-induced autophagy, BI-1-silenced TMV-infected NN plants accumulated more JOKA2 than nonsilenced control plants in either the presence (Fig. 4I) or the absence (Fig. S6G) of E64d. As it is reported that the transcriptional level of some ATG genes along with JOKA2 are elevated in different kinetics during the heat-stress-induced autophagy in plants,69 the transcriptional level of JOKA2 was tested by real-time PCR in this experiment, and the data showed that after TMV infection, the JOKA2 mRNA level in BI-1-silenced plants was downregulated compared with that in control plant either with or without E64d treatment (Fig. S6E and F). The above experiments suggest that silencing of BI-1 compromised autophagy flux and autophagy activity.
Taken together, these results suggest that plant BI-1 positively regulates the autophagy induced by N gene-mediated resistance to TMV.
Silencing of BI-1 compromised autophagy induced by MV
Methyl viologen (MV) is a reactive oxygen species (ROS) inducer in plants that can trigger autophagy.33,71 To test whether BI-1's role in autophagy is general, we used MV to trigger autophagy. CFP-ATG8F was used as a marker to label autophagic structures and transiently expressed followed by 10 µM MV treatment of 24 h before monitoring the fluorescence by confocal. BI-1-silenced plant showed decreased MV-induced autophagy in either the presence or absence of E64d as indicated by CFP-ATG8F-labeled autophagic structures (Fig. 4G, H, Fig. S6C and D). Further, the accumulation of JOKA2 in BI-1-silenced plant was more than that of nonsilenced plant in either the presence or absence of E64d (Fig. 4J, Fig. S6J). The mRNA level of JOKA2 was downregulated in BI-1-silenced plant after MV treatment in either the presence or absence of E64d (Fig. S6H and I), which was consistent with that in the TMV-induced autophagy experiment. These results suggest that silencing of BI-1 compromises the autophagy induced by MV. Therefore, plant BI-1 may have a general role in autophagy regulation.
Silencing of BI-1 enhanced N gene-mediated cell death
We previously found that ATG6 negatively regulated immunity-related PCD.10 We therefore evaluated the effect of silencing of BI-1 on N-mediated hypersensitive response (HR) PCD by TMV-GFP. BI-1-silenced plants had no developmental deficiency and showed no cell death (Fig. S7A). The HR was presented in all of the infected plants by 7 dpi with TMV-GFP. Furthermore, TMV-GFP accumulation was enhanced in BI-1-silenced NN plants compared with that in nonsilenced control NN plants, as observed by GFP fluorescence at the site of infection (Fig. 5A), and real-time RT-PCR (Fig. 5B). We also found enhanced cell death in BI-1-silenced plants (Fig. 5A, normal light). However, cell death did not spread beyond virus infection sites 15 dpi (Fig. S7B).
Figure 5.
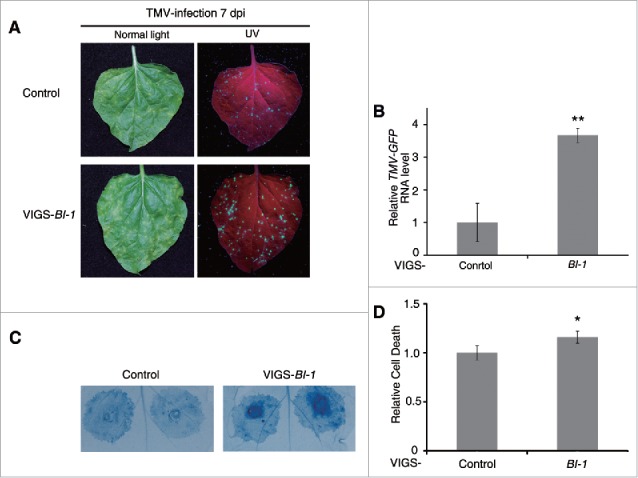
Silencing of BI-1 enhanced N-mediated cell death. (A) TMV-induced PCD was enhanced in BI-1-silenced N-containing plants. TMV-induced PCD was assessed in nonsilenced control (Control) and BI-1-silenced (VIGS-BI-1), N gene-containing plants. Representative images of TMV-GFP-infected leaves were taken under normal light and UV light at 7 dpi. Red represents chlorophyll autofluorescence. (B) Real-time RT-PCR showed relative RNA level of TMV-GFP in Control and VIGS-BI-1 plants. Leaves were collected 7 d post TMV-GFP infection. Data represent the mean ± SEM of 3 biological replicates with 4 technical replicates with mean value of control set to 1.0 (**P < 0.01, n = 12). Actin was used as an internal control. (C) Silencing of BI-1 enhanced TMV-p50 triggered cell death in N gene-containing plants. TMV-p50 was expressed by agroinfiltration in Control and VIGS-BI-1 plants. Trypan blue staining was performed after 48 hpi. (D) Quantitative representation of TMV-p50-induced HR programmed cell death. To quantify death intensity, images were converted to gray scale, and mean gray value of inoculation area was scored after subtracting the background of noninoculation area by ImageJ. Data represent means ± SEM (*P < 0.05, n = 64). Experiments were repeated 3 times.
Further, we found that the N gene-mediated cell death induced by the N gene elicitor, TMV-p50, also increased in BI-1-silenced plants, and cell death did not spread beyond the infiltrated regions either (Fig. 5C and D). To exclude the possibility of silencing of BI-1 on p50 expression, we test the TMV-p50 expression level in the BI-1-silenced nn plant and control nn plant, on which TMV-p50 would not cause HR PCD. The result showed that the protein level was similar (Fig. S7C), suggesting that the enhanced cell death in BI-1-silenced plant was not due to the different protein expression level of TMV-p50. These results suggest that tobacco BI-1 negatively regulates cell death during N-mediated resistance to TMV.
Overexpression of plant BI-1 induced autophagy and cell death in a dose-dependent manner
To test the effect of overexpression of plant BI-1 on autophagy, we expressed NbBI-1 in N. benthamiana by agroinfiltration. Overexpression of NbBI-1 driven by the CaMV 35S promoter with duplicated enhancers and TMV Omega leader sequence (2 × 35S-Ω:NbBI-1) caused macroscopic cell death by 3 dpi (data not shown). This result was surprising, since BI-1s had been reported to be the well-recognized cell death suppressors in plants and mammals.36,37,44,53 To confirm this observation, we expressed Arabidopsis BI-1 (AtBI-1) in N. benthamiana plants under the control of the CaMV 35S promoter (35S:AtBI-1), and CaMV 35S promoter with duplicated enhancers and a TMV Omega leader sequence (2 × 35S-Ω:AtBI-1). Accordingly, we also expressed cLUC under the control of 35S or 2 × 35S-Ω promoter respectively as negative controls (35S:cLUC and 2 × 35S-Ω:cLUC). The 2 × 35S version of the CaMV 35S promoter is reported to result in a tenfold higher transcriptional level than the natural CaMV 35S promoter (35S), while the inclusion of a TMV Omega leader sequence (Ω) also enhances the translational activity.72,73 Trypan blue staining indicated that regions agroinfiltrated with 35S:AtBI-1, 35S:cLUC or 2 × 35S-Ω:cLUC did not show any visible cell death by 3 dpi, whereas the areas agroinfiltrated with 2 × 35S-Ω:AtBI-1 displayed macroscopic cell death (Fig. 6A). These results suggest that plant BI-1 can have a prodeath role.
Figure 6.
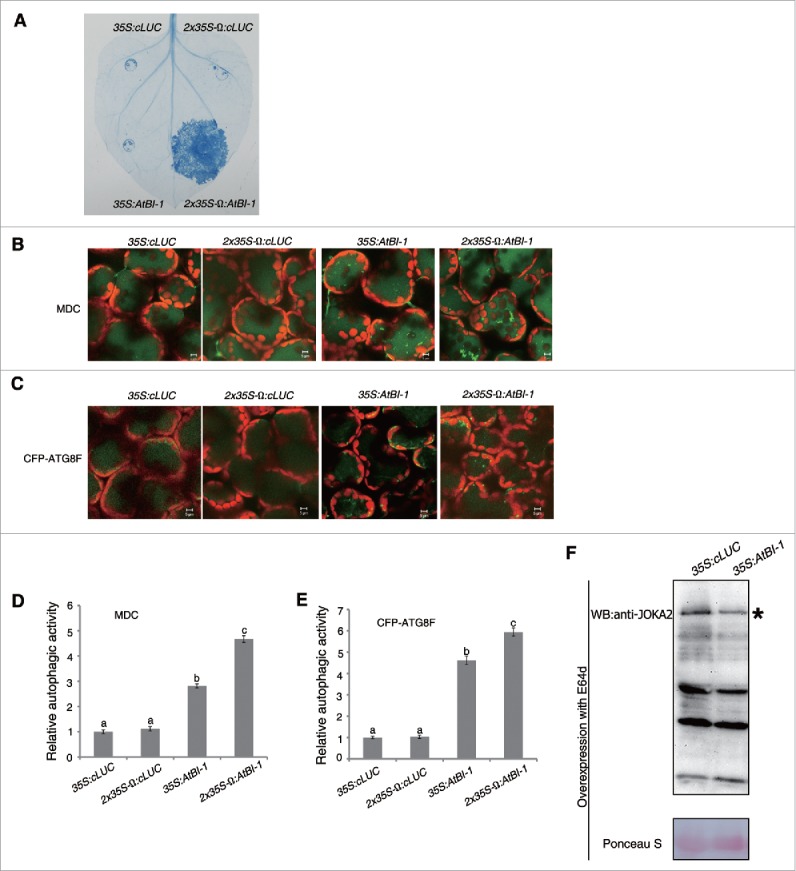
Overexpression of plant BI-1 trigged cell death and increased autophagic activity. (A) Trypan blue staining of cell death. Agrobacteria containing 35S:cLUC, 2 × 35S-Ω:cLUC, 35S:AtBI-1, and 2 × 35S-Ω:AtBI-1 were infiltrated into N. benthamiana. At 3 dpi, leaves were detached for trypan blue staining. (B) Representative images of MDC-stained autophagic structures from leaf tissues agroinfiltrated with 35S:cLUC, 2 × 35S-Ω:cLUC, 35S:AtBI-1, and 2 × 35S-Ω:AtBI-1 at 48 h postinfiltration (hpi), when no macroscopic cell death was apparent. (C) Representative images of CFP-ATG8F-labeled autophagic structures from leaf tissues agroinfiltrated with 35S:cLUC, 2 × 35S-Ω:cLUC, 35S:AtBI-1, and 2 × 35S-Ω:AtBI-1 at 48 hpi. The puncta in the mesophyll cells were autophagic structures. Scale bars ((B)and C): 5μm. (D) The relative autophagic activity in MDC-stained tissue. The number of MDC-stained autophagic structures per leaf section was counted and normalized to that of 35S:cLUC or 2 × 35S-Ω:cLUC control, respectively. Data are the means ± SEM of relative autophagic activity, with the mean value of control set to 1.0 (n = 10). Different letters (a to c) above bars represent significantly different groups. (E) The relative autophagic activity in CFP-ATG8F-labeled tissue. The number of CFP-ATG8F-labeled autophagic structures per leaf section was counted and normalized to control respectively. Data are the means ± SEM of relative autophagic activity, with the mean value of control set to 1.0 (n = 10). Different letters (a to c) above bars represent significantly different groups. (F) Overexpression of AtBI-1 decreased the accumulation of JOKA2. Ponceau S staining was used as loading control. Experiments were repeated at least 2 times. All autophagy-related experiments were performed with E64d treatment.
Whereas no signs of cell death were observed in the leaves of N. benthamiana plants agroinfiltrated with 2 × 35S:AtBI-1 or 2 × 35S-Ω:AtBI-1 at 48 h postinfiltration, we monitored the autophagic activity at this time point. We found much more MDC-stained autophagic structures in areas agroinfiltrated with 35S:AtBI-1 or 2 × 35S-Ω:AtBI-1 than in those agroinfiltrated with 35S:cLUC or 2 × 35S-Ω:cLUC (Fig. 6B and D) in the presence of E64d. Furthermore, MDC-stained structures were more in plants agroinfiltrated with 2 × 35S-Ω:AtBI-1 than those agroinfiltrated with 35S:AtBI-1 (Fig. 6B and D). No difference was found between 2 controls 35S:cLUC and 2 × 35S-Ω:cLUC (Fig. 6B and D). Consistent with MDC staining data, the number of CFP-ATG8F-labeled autophagic structures was greater in plants agroinfiltrated with 2 × 35S-Ω:AtBI-1 than those with 35S:AtBI-1 (Fig. 6C and E) in the presence of E64d. Further, we tested the autophagy activity by CFP-ATG8F assay in the absence of E64d, similar results were obtained (Fig. S8A and B). Also, accumulation of JOKA2 was decreased by overexpression of BI-1 in either the presence or absence of E64d (Fig. 6F, Fig. S8E). However, overexpression of BI-1 had no significant effect on JOKA2 mRNA level in either the presence or absence of E64d (Fig. S8C and D). Since overexpression of 2 × 35S-Ω:AtBI-1 caused cell death, which may affect general protein degradation process, the JOKA2 level was not tested in this condition. These results further suggest that plant BI-1 positively regulates autophagy.
To rule out the possibility that the increased cell death and autophagic activity were due to the overload of ER membrane protein, Arabidopsis RTNLB13 was used as a control. Arabidopsis RTNLB13, a member of the reticulon family, is an ER membrane protein and its overexpression remodels ER morphology, which was similar to the overexpression pattern of tobacco BI-1.74 The trypan blue staining result showed that expression of AtRTNLB13 driven by the 2 × 35S-Ω promoter could not cause cell death (Fig. S9B). Further, the level of CFP-ATG8F-labeled autophagic structures was similar in plants agroinfiltrated with 2 × 35S-Ω:cLUC and 2 × 35S-Ω:AtRTNLB13 (Fig. S9A). Taken together, these results suggest that BI-1 is able to specifically activate autophagy and induce cell death.
Autophagy was required for BI-1-mediated cell death
We also tested whether autophagy was required for BI-1-mediated cell death. To this end, we silenced ATG6, PI3K and ATG7, which are core autophagy genes.10 2 × 35S-Ω:AtBI-1 was transiently expressed in the silenced or nonsilenced control leaves. As expected, overexpression of AtBI-1 caused serious cell death in nonsilenced control plants (Fig. 7A). However, the cell death triggered by AtBI-1 was markedly suppressed in ATG6-, PI3K- or ATG7-silenced leaves (Fig. 7A). To address whether the protein level of AtBI-1 was comparable in nonsilenced and silenced plants, AtBI-1-YFP was generated under the control of the 2 × 35S-Ω promoter since the antibody against Arabidopsis BI-1 was unavailable. 2 × 35S-Ω:AtBI-1-YFP also caused cell death as 2 × 35S-Ω:AtBI-1, suggesting that YFP fused AtBI-1 was functional (Fig. S10A). The expression level of AtBI-1 was similar between ATG6-, PI3K- and ATG7-silenced plants and control plants 2 dpi, and the protein level was even lower in the control plant, which may be due to the potential cell death effect (Fig. S10B). Taken together these results suggest that autophagy is required for BI-1-mediated cell death.
Figure 7.
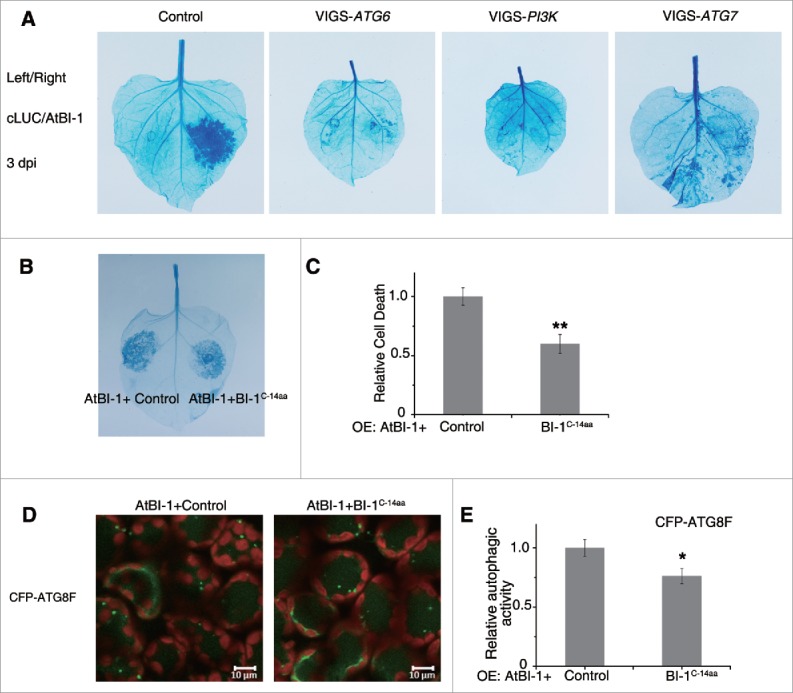
The prodeath role of plant BI-1 required the autophagy pathway. (A) Silencing of ATG6, PI3K or ATG7 suppressed the cell death induced by overexpression of AtBI-1. 2 × 35S-Ω:AtBI-1 was transiently expressed in the silenced leaves (VIGS-ATG6, VIGS-PI3K and VIGS-ATG7) or the nonsilenced control leaves (Control). At 3 dpi, leaves were detached for trypan blue staining. The experiment was repeated at least 3 times, using 5 or more plants per experiment. (B) Overexpression of C-terminal 14 aa of BI-1 decreases AtBI-1 triggered cell death. 2 × 35S-Ω:AtBI-1 was coexpressed with 35S:cLUC-3xHA (Control) or 35S:cLUC-NbBI-1C-14aa-HA (BI-1C-14aa) by agroinfiltration. Trypan blue staining was performed after 48 hpi. (C) Quantitative representation of AtBI-1-induced cell death. To quantify death intensity, images were converted to gray scale, and mean gray value of inoculation area was scored after subtracting the background of noninoculation area by ImageJ. Data represent means ± SEM (**P < 0.01, n = 6). The experiments were repeated 2 times. (D) Representative images of CFP-ATG8F-labeled autophagic structures from leaf tissues agroinfiltrated with 2 × 35S-Ω:AtBI-1, 35S:cLUC-3xHA (Control) or 35S:cLUC-NbBI-1C-14aa-HA (BI-1C-14aa) at 42 hpi. The puncta in the mesophyll cells were autophagic structures. (E) The relative autophagic activity in CFP-ATG8F-labeled tissue. The number of CFP-ATG8F-labeled autophagic structures per leaf section was counted and normalized to control. Data are the means±SEM of relative autophagic activity, with the mean value of the control set to 1.0 (*P < 0.05, n = 30). Experiments were repeated at least 2 times. All autophagy-related experiments were performed with E64d treatment.
The C-terminal 14 amino acids of BI-1 had effects on both BI-1-induced cell death and autophagy
Since the C-terminal 14 aa of BI-1 are conserved among species (Fig. S1), and are sufficient for the interaction between BI-1 and ATG6, we tested the function of the C-terminal 14 aa. C-terminal 14 aa of NbBI-1was tagged to cLUC along with a 1xHA on its C-terminal (cLUC-BI-1C-14aa-HA) for overexpression. When cLUC-BI-1C-14aa-HA (BI-1C-14aa) was expressed alone, no cell death was observed, and autophagy activity was not affected either. However, cell death induced by 2 × 35S-Ω:AtBI-1 decreased when coexpressed with BI-1C-14aa compared with that when coexpressed with control (cLUC-3xHA), as indicated by trypan blue staining (Fig. 7B and C). Western blot assay showed that when coexpressed with 2 × 35S-Ω:AtBI-1, the protein level of cLUC-3xHA were less than cLUC-BI-1C-14aa-HA (Fig. S10C), which may be due to the severe cell death in control cLUC-3xHA leaves.
Autophagy activity were measured using CFP-ATG8F assay. BI-1 induced autophagy were decreased when coexpressed with BI-1C-14aa compared with that when coexpressed with control in either the presence (Fig. 7D and E) or the absence (Fig. S10D and E) of E64d.
Taken together, the C-terminal 14 aa of BI-1 may interfere with the BI-1 induced cell death and autophagy.
Discussion
In this study, we found that plant BI-1, a novel ATG6-binding protein, positively regulates autophagy. Intriguingly, in addition to its well-recognized prosurvival role,44 plant BI-1 also has a prodeath role, and both of these activities are dependent on autophagy.
Roles of ATG6 in autophagy and cell death
ATG6/BECN1 is a phylogenetically conserved protein and is essential for the initiation of autophagy. Human BECN1 contains a BH3 domain, which is involved in the interaction with BCL2 or BCL2L1. Our study demonstrated the interaction of tobacco ATG6 with BI-1 in plants, and N-terminal domain of ATG6, which contains the putative BH3 domain, is sufficient for the interaction (Fig. 2). In mammals, the disruption of interaction between BECN1 and BCL2 or BCL2L increases the autophagic activity of BECN1.75 Further study to reveal the mechanism of BI-1's in autophagic activity could be focus on the ATG6's side. It will be interesting to test whether animal BI-1 interacts with ATG6 to competitively disrupt the interaction between ATG6 and BCL2 or BCL2L1 to regulate cell death since BCL2 or BCL2L1 orthologs have never been identified in plants. Unlike other known BH3-only proteins, ATG6 does not act as proapoptotic proteins in mammal. There exists crosstalk between cell death and autophagy by ATG6.76 It needs to be further investigated whether the interaction of plant ATG6 and BI-1 will have effect on BI-1 mediated cell death.
Roles of BI-1 in autophagy
Two recent reports describe the contrasting roles of animal BI-1 in autophagy. One report suggests that BI-1 arrests autophagy by inhibiting ERN1-mediated MAPK8 activation. When active, MAPK8 phosphorylates BCL2 to release ATG6 and promote autophagy.56 The second report suggests that BI-1 promotes autophagy in an ITPR-dependent manner. BI-1 could promote the ITPR-mediated release of ER Ca2+ into the cytosol, thus decreasing the transportation of Ca2+ from stores in the ER to mitochondria, which is necessary for the biosynthesis of ATP in mitochondria. The reduction in cellular ATP levels would in turn activate autophagy.57 The authors of the latter study proposes that the net outcome of these 2 distinct functions of BI-1 varies with the context.57 Here, we provide direct evidence that plant BI-1 interacts with the core autophagy component, ATG6, to promote autophagy. Silencing of BI-1 reduced autophagic activity induced by plant immune response and MV (Fig. 4). However, overexpression of Arabidopsis BI-1 increased autophagic activity (Fig. 6). In mammals, BECN1 interacts with ITPR to regulate autophagy.77 Recently, mammalian TMBIM6 has been reported to be a novel ITPR-interacting and -sensitizing protein.78 Our results demonstrate that ATG6 interacts directly with BI-1 in plant. It needs to be further investigated whether plant ITPR homologs can interact with both ATG6 and BI-1 to participate in autophagy. We assume that ER-localized BI-1 can interact with ATG6 to recruit the autophagy machinery components to initiate ER-associated autophagy.
Role of BI-1 and autophagy in cell death
BI-1 has been recognized as a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants.44,45 Numerous studies show that BI-1s are the conserved suppressors of cell death in different PCD pathways in both plants and animals. However, the underlying molecular mechanism was hitherto unclear. Like animal BI-1, plant BI-1s have been implicated in ER stress-induced PCD,46,52 and regulate cellular Ca2+ homeostasis by interacting with Arabidopsis AtCaM7 (Calmodulin 7).54,55 Furthermore, studies in plants suggest that BI-1 also modulates the sphingolipid-associated PCD pathway.79,80 In this study, we revealed that plant BI-1 interacts with ATG6 to promote autophagy and negatively regulates N gene-mediated PCD. We found that silencing of BI-1 decreased autophagic activity induced by N gene-mediated resistance to TMV and accelerated N-mediated cell death triggered by TMV elicitor p50, suggesting that the conserved prosurvival roles of BI-1 may rely on autophagy.
We demonstrated that plant BI-1 can function as a positive or negative regulator of cell death. Previous reports suggest that overexpression of Arabidopsis BI-1 in a human fibrosarcoma HT1080 cell line induces cell death.81,82 Our results showed that overaccumulation of BI-1 also led to HR-like cell death in plants. These observations suggest that plant BI-1 may also have a prodeath role in nature. Further, silencing of either ATG6, PI3K or ATG7 compromised BI-1-mediated cell death (Fig. 7A), suggesting that cell death triggered by overexpression of BI-1 requires autophagy. Similarly, immunity-associated cell death requires autophagy in Arabidopsis plants overexpressing a constitutively active form of RABG3B.32 Either silencing or knockout of GAPDHs in plants induces autophagy and enhances R-mediated cell death.33,83 Furthermore, Arabidopsis atg mutants exhibit a delay in HR cell death during the early stages of pathogen infection.30 Importantly, oxalic acid-deficient mutants of the necrotrophic phytopathogen Sclerotinia sclerotiorum trigger a restricted HR-like autophagic cell death.31 Additionally, autophagy has been suggested to promote death in tracheary element differentiation.84 These data suggest that excess autophagy is able to cause cell death and autophagy has a prodeath role in addition to its prosurvival role in plants.
It is reported that silencing or knockout of NbATG6 or other ATG genes causes the spreading of immunity-related cell death beyond the site of pathogen infection, describing the gradual spread of cell death to uninfected tissues in autophagy-deficient plants.10,29,85 However, N-mediated cell death did not spread to uninfected tissue in NbBI-1 silenced plants, although NbBI-1 is also required for autophagy induced by N-mediated resistance. Additionally, HR cell death mediated by the resistance protein, RPM1, does not spread beyond the infection site in younger atg mutants,30,85 but does in older atg mutants.85 Taken together, these observations suggest that HR cell death in autophagy-deficient mutants is uncoupled from spreading cell death. Given that the effect of autophagy on HR cell death at the primary infection site is controversial,23 it is possible that the effect of autophagy on HR PCD may promote either survival or death at the pathogen infection sites, depending on the host-pathogen system. However, autophagy normally antagonizes immunity-related cell death or enhanced senescence beyond the site of pathogen infection by alleviating ER stress and maintaining cellular homeostasis during aging.86 As the C-terminal 14 aa of BI-1 lowered BI-1-mediated cell death and autophagy (Fig. 7B to E), the overexpressed BI-1C-14aa could bind to endogenous ATG6 to weak the function of AtBI-1. Thus, the interaction between BI-1 and ATG6 is important for BI-1 induced autophagy and cell death.
Materials and methods
Yeast 2-hybrid assay and GST affinity isolation assay
Plasmids and yeast strains in yeast 2-hybrid assay were described and were used by following the instruction in the protocols (Dualmembrane system, P01501). GST affinity isolation assay was performed by following the kit's instructions (Thermo Fisher Scientific, 21516).
Luciferase complementation imaging (LCI) assay
LCI assays were performed as described.62 Generally, the plasmids were transformed into Agrobacterium GV3101 respectively. The agrobacterium cultures containing the pairs of plasmids used to test for interactions were cultured, harvested and resuspended in buffer with 10 mM MgCl2 [Sinopharm Chemical Reagent, 10012818], 10 mM MES [AMRESCO, E169], and 200 μM acetosyringone [Sigma-Aldrich, D134406]. Bacterial suspensions were infiltrated into N. benthamiana leaves by needleless syringe. 1 μM luciferin (Gold Biotechnology, LUCK-1G) was sprayed on to leaves after 48 h infiltration before relative LUC activity test. Luminescence was captured with a low-light cooled Andor iXon CCD camera (Andor Technology, South Windsor, CT, USA) after a 5 min exposure.
Coimmunoprecipitation (CoIP) and western blot assays
Total proteins from leaves were extracted with a ratio of 1:1 of native extraction buffer (NB buffer) (50 mM TRIS-MES [TRIS AMRESCO, 0497; MES AMRESCO, E169], pH 8.0, 0.5 M sucrose [Beijing Chemical Works, K0800043], 1 mM MgCl2 [Sinopharm Chemical Reagent, 10012818], 10 mM EDTA [Sinopharm Chemical Reagent, 10009717], 10 mM DTT [Sigma-Aldrich, D0632], protease inhibitor cocktail CompleteMini tablets [Roche, 04693124001], phosphatase inhibitor [Roche, PHOSS-RO]).87 Protein extracts were incubated with the Anti-MYC-Tag mAb beads (Abmart, M20012) for 2 h at 4°C, and the beads were washed 3 times with ice-cold NB buffer at 4°C. IP samples were analyzed by SDS-PAGE, immunoblotted using anti-HA (Cell Signaling Technology, 2999) and anti-MYC antibodies (Abmart, 19C2) and detected using Pierce ECL western blotting substrate (Thermo Fisher Scientific, 32209).
In NBR1/JOKA2 western blot assay, plant leaves were harvested and grinded by liquid nitrogen and added 5% 2-Mercaptoethanol (Sigma-Aldrich, M3148–100ML) 2 × loading buffer (100 mg: 200 μL). Anti-NBR1 (Agrisera, AS14 2805) antibody was used to test the endogenous Joka2 of tobacco.
Confocal microscopy and transmission electron microscopy
In BiFC assay, the plasmids were transformed into Agrobacterium GV3101. The agrobacterium cultures containing the pairs of plasmids used to test for interactions between proteins and the ER marker or Golgi marker G-cb were mixed at a ratio of 1:1:1 and were used to coinoculate N. benthamiana plants.60 After 48 h of expression, the injected area was punched out using a 0.6-cm diameter cork borer and was observed using an inverted Zeiss LSM 710 laser scanning microscope (Carl Zeiss, Jena, Germany) (www.zeiss.com) and a Plan-Apochromat 40x/0.95 Korr M27 water immersion objective (Carl Zeiss, Jena, Germany). For CFP and YFP imaging, the emission wavelengths were excited using the LD laser line at 405 nm (CFP) and the Multi Ar laser lines at 514 nm (YFP) respectively. The frame-scanning mode was applied for the acquisition of CFP and YFP signals.
The MDC staining assay was performed as described;28 the leaves were infiltrated with 100 μM E64d (Sigma-Aldrich, E8640) and treated in the dark for 10 to 12 h. After the dark treatment, the E64d-infiltrated parts of the leaves were excised and immediately vacuum infiltrated with 50 μM MDC (Sigma-Aldrich, 30432) for 10 min, followed by 2 washes with phosphate-buffered saline buffer pH 7.4 (137 mM NaCl [Sinopharm Chemical Reagent, 10019308], 2.7 mM KCl [Sinopharm Chemical Reagent, 10016308], 10 mM Na2HPO4 [Sinopharm Chemical Reagent, 20040618], 1.8 mM KH2PO4 [Sinopharm Chemical Reagent, 10017608]). MDC-incorporated structures were excited by a wavelength of 405 nm and detected at 400 to 580 nm. Chloroplast autofluorescence was excited at 543 nm and detected at 580 to 695 nm.
When using the autophagy marker CFP-ATG8F,33 Agrobacterium with CFP-ATG8F plasmid was infiltrated into N. benthamiana leaves. Two d after agroinfiltration, the samples underwent additional infiltration with 100 μM E64d for 10–12 h. Leaves were then excised and incubated in phosphate-buffered saline buffer, then autophagosomes were monitored using a Zeiss LSM 710 at 405 nm. The localization of BI-1-YFP was monitored by a 514-nm excitation laser. Experiments were performed as described previously.
MV (10 µM; Sigma-Aldrich, STBB4829) was inoculated on leaves 24 h before monitoring the fluorescence by confocal.
VIGS assay, virus infection, and GFP imaging
Experiments were performed as described previously.10 Agrobacteria mixture containing pTRV1 and pTRV2 or pTRV2 derivative plasmids was grown overnight at 28°C respectively. Bacteria were resuspended and mixed at 1:1 ratio. After 4 h incubation at room temperature, the mixed Agrobacterium cultures were infiltrated into the leaves of 6-leaf stage N. benthamiana plants. Silenced phenotypes appeared in the upper leaves ∼10 d postinfiltration. NN plants were used for TMV-GFP and TMV-p50 helicase assays. The upper leaves of the VIGS-BI-1 plants were infected with TMV-GFP. GFP imaging was performed using UV illumination and photographs were taken by digital camera.
Statistical analysis
The data were expressed as mean ± SEM if there is no specific statement otherwise. Statistical analysis was performed using the Student t test with P = >0.05 not significant (NS), *P = <0.05, **P = <0.01, ***P = <0.001. The Levene test of homogeneity of variance was applied before ANOVA. One-way ANOVA followed by the LSD-t test was used for statistical analysis when equal variances were assumed (P value of the Levene test was greater than or equal to 0.05), and one-way ANOVA followed by the Tamhane T2 test was used for statistical analysis when equal variances were not assumed (P value of the Levene test was less than 0.05). Different letters (a to c) above bars represent significantly different groups.
Supplementary Material
Abbreviations
- At
Arabidopsis thaliana
- ATG
autophagy related
- BAX
BCL2 associated X, apoptosis regulator
- BCL2, BCL2
apoptosis regulator
- BCL2L1/BCL-XL
BCL2 like 1
- BI-1
Bax inhibitor-1
- BiFC
bimolecular fluorescence complementation
- cLUC
C-terminal of luciferase
- DPI
days postinfiltration
- ER
endoplasmic reticulum
- ERN1/IRE1
endoplasmic reticulum to nucleus signaling 1
- GST
glutathione S-transferase
- HPI
hours postinfiltration
- HR
hypersensitive response
- ITPR
inositol 1,4,5-trisphosphate receptor
- LUC
luciferase
- MAPK8/JNK1
mitogen-activated protein kinase 8
- MDC
monodansylcadaverine
- MV
methyl viologen
- Nb
Nicotiana benthamiana
- Nt
Nicotiana tabacum
- nLUC
N-terminal of luciferase
- PCD
programmed cell death
- PtdIns3K
phosphatidylinositol 3-kinase
- TEM
transmission electron microscopy
- TMBIM6
transmembrane BAX inhibitor motif containing 6
- TMV
Tobacco mosaic virus
- TRV
Tobacco rattle virus
- VIGS
virus-induced gene silencing.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ying Li at the Center of Biomedical Analysis, Tsinghua University for helping with the TEM and confocal assays. Thank Prof. Martin Dickman at Texas A & M University and Prof. Xinquan Wang at Tsinghua University for the helps in ATG6 structure prediction. Thanks to Oliver. M. Terrett for polishing the manuscripts.
Funding
This work was supported by the National Natural Science Foundation of China (31530059, 31421001, 31470254, 31300218 and 31270182), the National Basic Research Program of China (2014CB138400).
References
- [1].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124-31; PMID:20034776; https://doi.org/ 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Bio 2007; 8:931-7; https://doi.org/ 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- [3].Il Kwon S, Park OK. Autophagy in Plants. J Plant Biol 2008; 51:313-20; https://doi.org/ 10.1007/BF03036132 [DOI] [Google Scholar]
- [4].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; https://doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Orsi A, Polson HEJ, Tooze SA. Membrane trafficking events that partake in autophagy. Curr Opin Cell Biol 2010; 22:150-6; PMID:20036114; https://doi.org/ 10.1016/j.ceb.2009.11.013 [DOI] [PubMed] [Google Scholar]
- [6].Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ 2005; 12:1542-52; PMID:16247502; https://doi.org/ 10.1038/sj.cdd.4401765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Oberstein A, Jeffrey PD, Shi YG. Crystal structure of the Bcl-X-L-beclin 1 peptide complex - Beclin 1 is a novel BH3-only protein. J Biol Chem 2007; 282:13123-32; PMID:17337444; https://doi.org/ 10.1074/jbc.M700492200 [DOI] [PubMed] [Google Scholar]
- [8].Funderburk SF, Wang QJ, Yue Z. The Beclin 1–VPS34 complex – at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355-62; PMID:20356743; https://doi.org/ 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Welters P, Takegawa K, Emr SD, Chrispeels MJ. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci U S A 1994; 91:11398-402; PMID:7972072; https://doi.org/ 10.1073/pnas.91.24.11398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005; 121:567-77; PMID:15907470; https://doi.org/ 10.1016/j.cell.2005.03.007 [DOI] [PubMed] [Google Scholar]
- [11].He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140-9; PMID:20097051; https://doi.org/ 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Y, Zheng X, Yu B, Han S, Guo J, Tang H, Yu AY, Deng H, Hong Y, Liu Y. Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy 2015; 11:2259-74; PMID:26566764; https://doi.org/ 10.1080/15548627.2015.1113365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 2008; 4:600-6; https://doi.org/ 10.4161/auto.6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol 2007; 372:223-35; PMID:17659302; https://doi.org/ 10.1016/j.jmb.2007.06.069 [DOI] [PubMed] [Google Scholar]
- [15].Ku B, Woo JS, Liang C, Lee KH, Hong HS, Xiaofei E, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS pathogens 2008; 4:e25; PMID:18248095; https://doi.org/ 10.1371/journal.ppat.0040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maiuri MC, Le Toumelin G, Criollo A, Rain J-C, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al.. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J 2007; 26:2527-39; PMID:17446862; https://doi.org/ 10.1038/sj.emboj.7601689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem 2009; 284:2719-28; PMID:19029119; https://doi.org/ 10.1074/jbc.M805920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 2009; 5:720-2; PMID:19395874; https://doi.org/ 10.4161/auto.5.5.8625 [DOI] [PubMed] [Google Scholar]
- [19].Lindqvist LM, Heinlein M, Huang DCS, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. P Natl Acad Sci USA 2014; 111:8512-7; https://doi.org/ 10.1073/pnas.1406425111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Y, Kabbage M, Liu W, Dickman MB. Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants. Plant Cell 2016; 28:233-47; PMID:26739014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haxim Y, Ismayil A, Jia Q, Wang Y, Zheng X, Chen T, Qian L, Liu N, Wang Y, Han S, et al.. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017; 6:e23897; PMID:28244873; https://doi.org/ 10.7554/eLife.23897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol 2007; 10:587-93; PMID:17702643; https://doi.org/ 10.1016/j.pbi.2007.06.006 [DOI] [PubMed] [Google Scholar]
- [23].Han S, Yu B, Wang Y, Liu Y. Role of plant autophagy in stress response. Protein Cell 2011; 2:784-91; PMID:22058033; https://doi.org/ 10.1007/s13238-011-1104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012; 24:4635-51; PMID:23175745; https://doi.org/ 10.1105/tpc.112.101535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 2002; 277:33105-14; PMID:12070171; https://doi.org/ 10.1074/jbc.M204630200 [DOI] [PubMed] [Google Scholar]
- [26].Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 2002; 129:1181-93; PMID:12114572; https://doi.org/ 10.1104/pp.011024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009; 5:954-63; PMID:19587533; https://doi.org/ 10.4161/auto.5.7.9290 [DOI] [PubMed] [Google Scholar]
- [28].Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al.. Autophagy contributes to leaf starch degradation. Plant Cell 2013; 25:1383-99; PMID:23564204; https://doi.org/ 10.1105/tpc.112.108993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 2008; 4:20-7; PMID:17932459; https://doi.org/ 10.4161/auto.5056 [DOI] [PubMed] [Google Scholar]
- [30].Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, et al.. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 2009; 137:773-83; PMID:19450522; https://doi.org/ 10.1016/j.cell.2009.02.036 [DOI] [PubMed] [Google Scholar]
- [31].Kabbage M, Williams B, Dickman MB. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS pathogens 2013; 9:e1003287; PMID:23592997; https://doi.org/ 10.1371/journal.ppat.1003287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kwon SI, Cho HJ, Kim SR, Park OK. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol 2013; 161:1722-36; PMID:23404918; https://doi.org/ 10.1104/pp.112.208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y. Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell 2015; 27:1316-31; PMID:25829441; https://doi.org/ 10.1105/tpc.114.134692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell 1998; 1:337-46; PMID:9660918; https://doi.org/ 10.1016/S1097-2765(00)80034-9 [DOI] [PubMed] [Google Scholar]
- [35].Szegezdi E, MacDonald DC, Ní Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol 2009; 296:C941-C53; PMID:19279228; https://doi.org/ 10.1152/ajpcell.00612.2008 [DOI] [PubMed] [Google Scholar]
- [36].Chae HJ, Ke N, Kim HR, Chen S, Godzik A, Dickman M, Reed JC. Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 2003; 323:101-13; PMID:14659883; https://doi.org/ 10.1016/j.gene.2003.09.011 [DOI] [PubMed] [Google Scholar]
- [37].Huckelhoven R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 2004; 9:299-307; PMID:15258461; https://doi.org/ 10.1023/B:APPT.0000025806.71000.1c [DOI] [PubMed] [Google Scholar]
- [38].Westphalen BC, Wessig J, Leypoldt F, Arnold S, Methner A. BI-1 protects cells from oxygen glucose deprivation by reducing the calcium content of the endoplasmic reticulum. Cell Death Differ 2005; 12:304-6; PMID:15650756; https://doi.org/ 10.1038/sj.cdd.4401547 [DOI] [PubMed] [Google Scholar]
- [39].Kim H-R, Lee G-H, Ha K-C, Ahn T, Moon J-Y, Lee B-J, Cho SG, Kim S, Seo YR, Shin YJ, et al.. Bax Inhibitor-1 Is a pH-dependent Regulator of Ca2+ Channel Activity in the Endoplasmic Reticulum. J Biol Chem 2008; 283:15946-55; PMID:18378668; https://doi.org/ 10.1074/jbc.M800075200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu C, Xu W, Palmer AE, Reed JC. BI-1 Regulates Endoplasmic Reticulum Ca2+ Homeostasis Downstream of Bcl-2 Family Proteins. J Biol Chem 2008; 283:11477-84; PMID:18299329; https://doi.org/ 10.1074/jbc.M708385200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al.. BAX Inhibitor-1 Is a Negative Regulator of the ER Stress Sensor IRE1 alpha. Mol Cell 2009; 33:679-91; PMID:19328063; https://doi.org/ 10.1016/j.molcel.2009.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Robinson KS, Clements A, Williams AC, Berger CN, Frankel G. Bax inhibitor 1 in apoptosis and disease. Oncogene 2011; 30:2391-400; PMID:21297665; https://doi.org/ 10.1038/onc.2010.636 [DOI] [PubMed] [Google Scholar]
- [43].Yang X, Srivastava R, Howell SH, Bassham DC. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J 2016; 85:83-95; PMID:26616142; https://doi.org/ 10.1111/tpj.13091 [DOI] [PubMed] [Google Scholar]
- [44].Watanabe N, Lam E. Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci 2009; 10:3149-67; PMID:19742129; https://doi.org/ 10.3390/ijms10073149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Watanabe N, Lam E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J 2006; 45:884-94; PMID:16507080; https://doi.org/ 10.1111/j.1365-313X.2006.02654.x [DOI] [PubMed] [Google Scholar]
- [46].Duan YF, Zhang WS, Li B, Wang YN, Li KX, Sodmergen, Han C, Zhang Y, Li X. An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol 2010; 186:681-95; PMID:20298483; https://doi.org/ 10.1111/j.1469-8137.2010.03207.x [DOI] [PubMed] [Google Scholar]
- [47].Babaeizad V, Imani J, Kogel KH, Eichmann R, Huckelhoven R. Over-expression of the cell death regulator BAX inhibitor-1 in barley confers reduced or enhanced susceptibility to distinct fungal pathogens. Theor Appl Genet 2009; 118:455-63; PMID:18956174; https://doi.org/ 10.1007/s00122-008-0912-2 [DOI] [PubMed] [Google Scholar]
- [48].Kawai-Yamada M, Jin L, Yoshinaga K, Hirata A, Uchimiya H. Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc Natl Acad Sci USA 2001; 98:12295-300; PMID:11593047; https://doi.org/ 10.1073/pnas.211423998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Eichmann R, Schultheiss H, Kogel KH, Huckelhoven R. The barley apoptosis suppressor homologue BAX inhibitor-1 compromises nonhost penetration resistance of barley to the inappropriate pathogen Blumeria graminis f. sp. tritici. Mol Plant Microbe Interact 2004; 17:484-90; PMID:15141952; https://doi.org/ 10.1094/MPMI.2004.17.5.484 [DOI] [PubMed] [Google Scholar]
- [50].Eichmann R, Dechert C, Kogel KH, Huckelhoven R. Transient over-expression of barley BAX Inhibitor-1 weakens oxidative defence and MLA12-mediated resistance to Blumeria graminis f.sp. hordei. Mol Plant Pathol 2006; 7:543-52; PMID:20507468; https://doi.org/ 10.1111/j.1364-3703.2006.00359.x [DOI] [PubMed] [Google Scholar]
- [51].Sanchez P, De Torres Zabala M, Grant M. AtBI-1, a plant homologue of Bax Inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J 2000; 21:393-9; PMID:10758491; https://doi.org/ 10.1046/j.1365-313x.2000.00690.x [DOI] [PubMed] [Google Scholar]
- [52].Watanabe N, Lam E. BAX Inhibitor-1 Modulates Endoplasmic Reticulum Stress-mediated Programmed Cell Death in Arabidopsis. J Biol Chem 2008; 283:3200-10; PMID:18039663; https://doi.org/ 10.1074/jbc.M706659200 [DOI] [PubMed] [Google Scholar]
- [53].Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ 2011; 18:1271-8; PMID:21597463; https://doi.org/ 10.1038/cdd.2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kawai-Yamada M, Hori Z, Ogawa T, Ihara-Ohori Y, Tamura K, Nagano M, Ishikawa T, Uchimiya H. Loss of calmodulin binding to Bax inhibitor-1 affects Pseudomonas-mediated hypersensitive response-associated cell death in Arabidopsis thaliana. J Biol Chem 2009; 284:27998-8003; PMID:19674971; https://doi.org/ 10.1074/jbc.M109.037234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ihara-Ohori Y, Nagano M, Muto S, Uchimiya H, Kawai-Yamada M. Cell death suppressor Arabidopsis bax inhibitor-1 is associated with calmodulin binding and ion homeostasis. Plant Physiol 2007; 143:650-60; PMID:17142482; https://doi.org/ 10.1104/pp.106.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Castillo K, Rojas-Rivera D, Lisbona F, Caballero B, Nassif M, Court FA, Schuck S, Ibar C, Walter P, Sierralta J, et al.. BAX inhibitor-1 regulates autophagy by controlling the IRE1alpha branch of the unfolded protein response. EMBO J 2011; 30:4465-78; PMID:21926971; https://doi.org/ 10.1038/emboj.2011.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sano R, Hou Y-CC, Hedvat M, Correa RG, Shu C-W, Krajewska M, Diaz PW, Tamble CM, Quarato G, Gottlieb RA, et al.. Endoplasmic reticulum protein BI-1 regulates Ca2+-mediated bioenergetics to promote autophagy. Genes Dev 2012; 26:1041-54; PMID:22588718; https://doi.org/ 10.1101/gad.184325.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, et al.. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res 2012; 22:473-89; PMID:22310240; https://doi.org/ 10.1038/cr.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cebulski J, Malouin J, Pinches N, Cascio V, Austriaco N. Yeast Bax inhibitor, Bxi1p, is an ER-localized protein that links the unfolded protein response and programmed cell death in Saccharomyces cerevisiae. Plos One 2011; 6:e20882; PMID:21673967; https://doi.org/ 10.1371/journal.pone.0020882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 2007; 51:1126-36; PMID:17666025; https://doi.org/ 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- [61].Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998; 72:8586-96; PMID:9765397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 2008; 146:368-76; PMID:18065554; https://doi.org/ 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 2007; 5:e68; PMID:17298188; https://doi.org/ 10.1371/journal.pbio.0050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J 2002; 31:777-86; PMID:12220268; https://doi.org/ 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- [65].Mitou G, Budak H, Gozuacik D. Techniques to study autophagy in plants. Int J Plant Genomics 2009; 2009:451357; PMID:19730746; https://doi.org/ 10.1155/2009/451357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 2009; 149:220-34; PMID:18790996; https://doi.org/ 10.1104/pp.108.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al.. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505-16; PMID:19250911; https://doi.org/ 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- [68].Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzyńska A, Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011; 7:1145-58; PMID:21670587; https://doi.org/ 10.4161/auto.7.10.16617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet 2013; 9:e1003196; PMID:23341779; https://doi.org/ 10.1371/journal.pgen.1003196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hackenberg T, Juul T, Auzina A, Gwizdz S, Malolepszy A, Van Der Kelen K, Dam S, Bressendorff S, Lorentzen A, Roepstorff P, et al.. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 2013; 25:4616-26; PMID:24285797; https://doi.org/ 10.1105/tpc.113.117192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 2007; 143:291-9; PMID:17098847; https://doi.org/ 10.1104/pp.106.092106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gallie DR, Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res 1992; 20:4631-8; PMID:1408765; https://doi.org/ 10.1093/nar/20.17.4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 1987; 236:1299-302; PMID:17770331; https://doi.org/ 10.1126/science.236.4806.1299 [DOI] [PubMed] [Google Scholar]
- [74].Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 2008; 9:94-102; PMID:17980018; https://doi.org/ 10.1111/j.1600-0854.2007.00670.x [DOI] [PubMed] [Google Scholar]
- [75].Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al.. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26:2527-39; PMID:17446862; https://doi.org/ 10.1038/sj.emboj.7601689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18:571-80; PMID:21311563; https://doi.org/ 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vicencio JM, Ortiz C, Criollo A, Jones AWE, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al.. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 2009; 16:1006-17; PMID:19325567; https://doi.org/ 10.1038/cdd.2009.34 [DOI] [PubMed] [Google Scholar]
- [78].Kiviluoto S, Schneider L, Luyten T, Vervliet T, Missiaen L, De Smedt H, Parys JB, Methner A, Bultynck G. Bax Inhibitor-1 is a novel IP3 receptor-interacting and -sensitizing protein. Cell Death Dis 2012; 3:e367; PMID:22875004; https://doi.org/ 10.1038/cddis.2012.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nagano M, Ihara-Ohori Y, Imai H, Inada N, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b(5). Plant J 2009; 58:122-34; PMID:19054355; https://doi.org/ 10.1111/j.1365-313X.2008.03765.x [DOI] [PubMed] [Google Scholar]
- [80].Nagano M, Takahara K, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. Arabidopsis Sphingolipid Fatty Acid 2-Hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-Hydroxylation and stress responses. Plant Physiol 2012; 159:1138-48; PMID:22635113; https://doi.org/ 10.1104/pp.112.199547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yu LH, Kawai-Yamada M, Naito M, Watanabe K, Reed JC, Uchimiya H. Induction of mammalian cell death by a plant Bax inhibitor. FEBS Letters 2002; 512:308-12; PMID:11852101; https://doi.org/ 10.1016/S0014-5793(02)02230-5 [DOI] [PubMed] [Google Scholar]
- [82].Lam E. Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 2004; 5:305-15; PMID:15071555 [DOI] [PubMed] [Google Scholar]
- [83].Henry E, Fung N, Liu J, Drakakaki G, Coaker G. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet 2015; 11:e1005199; PMID:25918875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kwon SI, Cho HJ, Park OK. Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation. Autophagy 2010; 6:1187-9; PMID:20861670 [DOI] [PubMed] [Google Scholar]
- [85].Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009; 21:2914-27; PMID:19773385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Munch D, Rodriguez E, Bressendorff S, Park O, Hofius D, Petersen M. Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy 2014; 10:54-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jia Q, Liu N, Xie K, Dai Y, Han S, Zhao X, Qian L, Wang Y, Zhao J, Gorovits R, et al.. CLCuMuB betaC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathogens 2016; 12:e1005668; PMID:27315204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


