Figure 3.
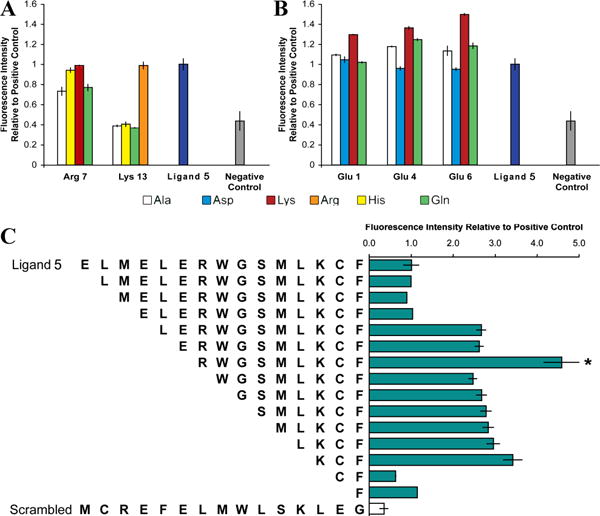
Identification of key residues, and removal of detrimental amino acids. a) In the library of similar and dissimilar substitutions of amino acids in ligand 5, at Arg7, ligand binding was retained for Lys and His substituents but was abolished by neutral Gln or Ala substituents. At Lys13, ligand binding was eliminated for all substituents except Arg. We conclude that both these residues contribute to binding primarily via positive charge. b) For Glu1, ligand binding was retained for negatively charged Asp and neutral Gln, and was slightly elevated for Ala. For Glu4 and Glu6, ligand binding increased moderately for neutral Gln and Ala. In short, positively charged residues are optimal, followed by neutral residues. Ligand binding was reduced by substitution with negatively charged Asp, and all Lys substitutions at these three sites increased ligand binding. These trends suggest that these Glu are not optimal for binding. c) Truncation of the six N-terminal residues, which included all three Glu, without removing any of the positively charged residues, yielded a peptide (indicated by asterisk) designated ligand 6 that became the template for subsequent library design. All libraries include a scrambled ligand 5 sequence as a negative control.
