Figure 2.
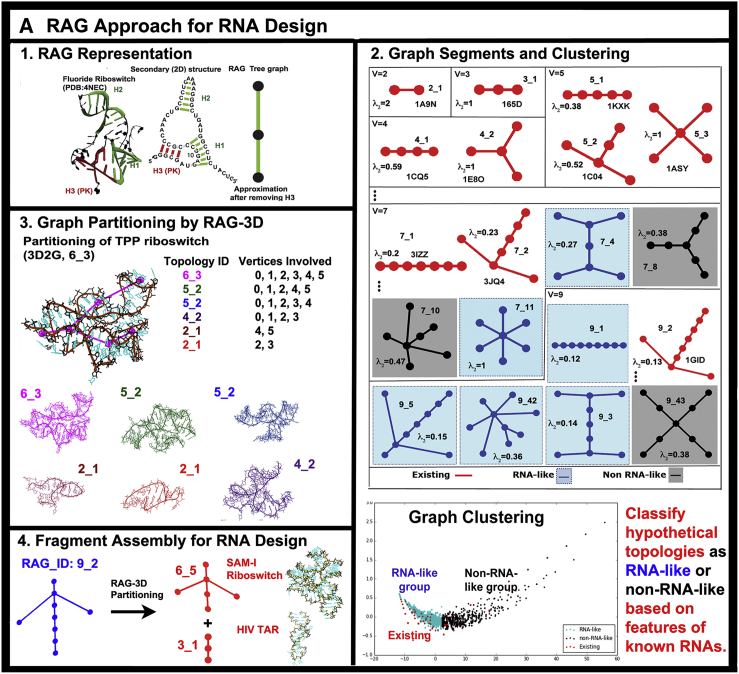
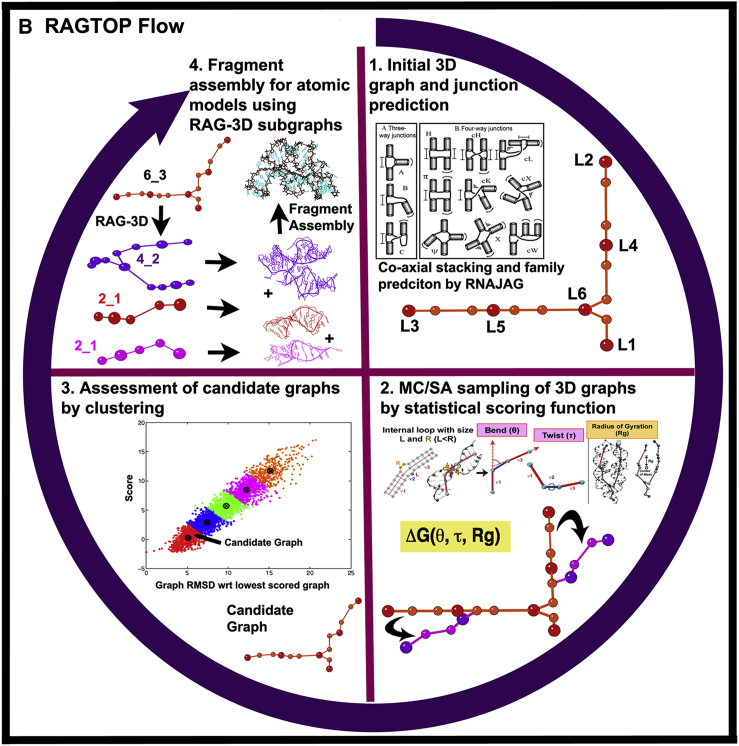
RAG elements for RNA motif classification, prediction, partitioning, and design. (A) The RAG approach for RNA representation (42) and design (43, 45) is illustrated as: 1) tree graph of a riboswitch; 2) RAG tree graph catalog segments, organized by the second eigenvalue λ2 of the connectivity matrix (Laplacian) associated with the graph and classified by clustering into three groups: existing (red), RNA-like (blue), and non-RNA-like (black) motifs (see http://www.biomath.nyu.edu/rag/home for more information); 3) graph partitioning for a riboswitch by RAG-3D (44), which suggests modular RNA building blocks, for which PDB structures are available; and 4) fragment assembly of such subgraph fragments using the modular subunits. (B) RAGTOP for 3D structure prediction by a hierarchical MC sampling of tree graphs (46, 48, 49): 1) Initial junction topology prediction; 2) MC sampling of 3D graphs scored by a statistical scoring function with components for bend, twist, and radius-of-gyration; 3) clustering of generated graphs to identify candidate graph; and 4) determination of atomic models from the candidate graph using the fragment assembly based on RAG-3D subgraphs. To see this figure in color, go online.