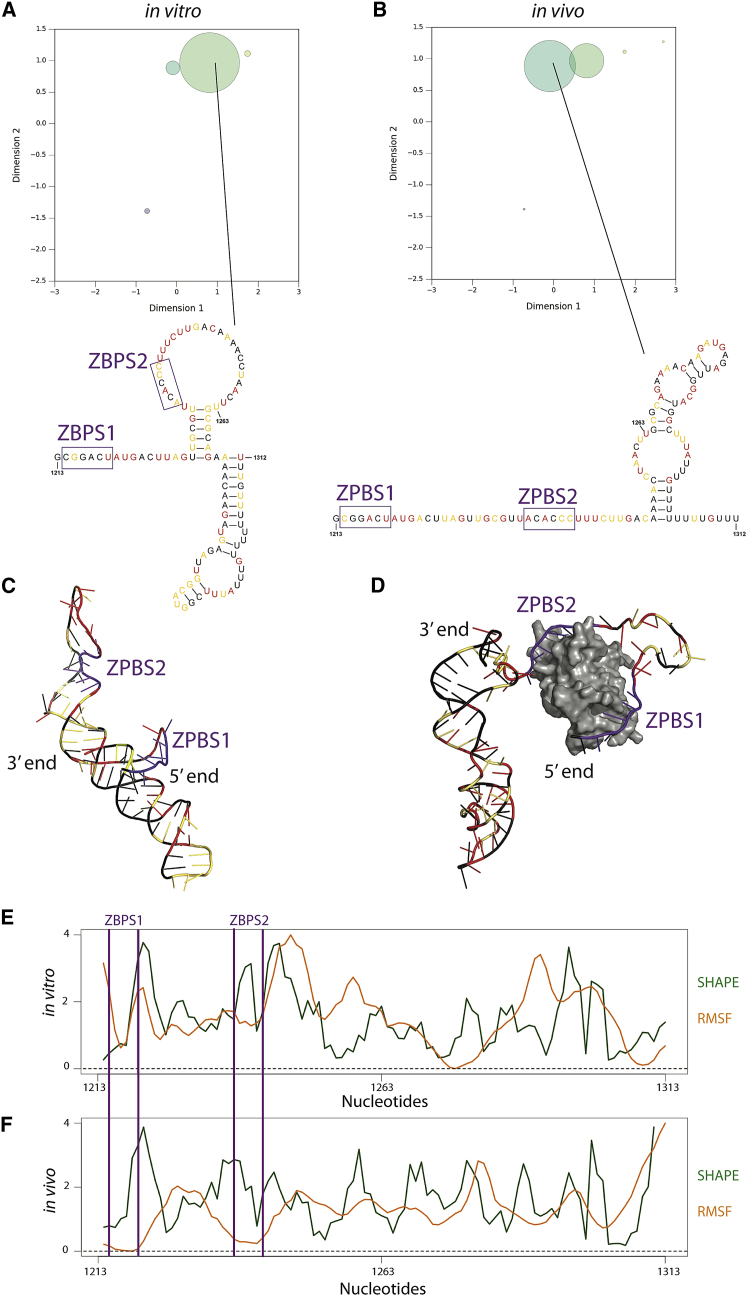
Figure 5.
Ensemble visualization for in vitro and in vivo human β-actin mRNA. Generation of structures for the β-actin mRNA ensemble was guided by the in vitro and in vivo SHAPE data. We compared the (A) in vitro and (B) in vivo ensembles for the region where SHAPE reactivities were expected to be different. The ensemble visualization reveals a large shift away from the dominant structure in vitro toward a second structure in vivo. We visualized the second structure for the medoid in each of the largest structure clusters. These nucleotides form different structures in vitro and in vivo. The region that differs includes the zipcode region with the two ZBP1 binding sites (purple). (C) The 3D structure for β-actin in vitro was modeled using molecular dynamics simulations without ZBP1. (D) The 3D structure for β-actin in vivo was modeled with the ZBP1 (in gray). For both 3D models, the ZBP1 binding regions are highlighted in purple. Red nucleotides correspond to high SHAPE reactivity, yellow correspond to medium reactivity, and black corresponds to low reactivity in (A–D) and (E). Comparison of SHAPE reactivity (green) and normalized RMSF (orange) for β-actin in vitro largely follow the same pattern. (F) Comparison of SHAPE reactivity and RMSF for β-actin in vivo also largely follow the same pattern. The SHAPE reactivities and RMSF values are averaged across a 3-nucleotide moving window. The RMSF is calculated from the 3D structural models. ZPB1 binding sites for (E) and (F) are boxed in purple. Fig. S6 includes further comparisons between in vitro and in vivo SHAPE reactivity and RMSF. To see this figure in color, go online.