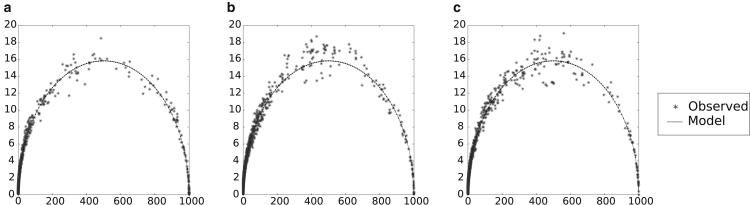
Figure 1.
Actual versus model standard deviation for helix classes of (a) Haloferax volcanii, (b) Escherichia coli, and (c) Encephalitozoon cuniculi 16S rRNA sequences. These sequences have been shown to have very different MFE accuracies and behaviors under SHAPE perturbation (29); their helix-class frequency behaviors, however, are seen to be similar, and thus are assumed to be typical. One hundred samples of 1000 structures each were generated for the sequences, using the same unperturbed, original set of parameters. To gauge the normal level of helix-class frequency variation, the standard deviation for each helix class frequency was calculated (i.e., the square root of the average of the squared deviations from the mean). Dots represent a helix class, with the mean, μ, of its frequency across 100 samples as its x coordinate, and the calculated standard deviation, , of its frequency across 100 samples as its y coordinate. The curve represents the model standard deviation, calculated as , where p is the ratio of the observed frequency of the helix class over the sample size, n. In general, a very good agreement exists between actual and model standard deviations.