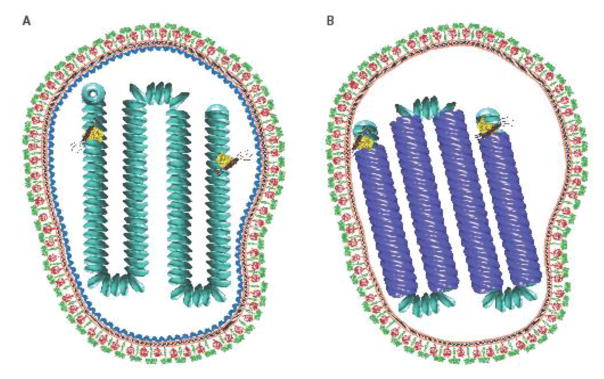
Figure 1.
A) Model of an MeV virion in which the matrix protein coats the nucleocapsid protein. The viral envelope is shown in orange. The nucleocapsid proteins are shown in cyan. The fusion protein trimers are shown in red. The attachment glycoprotein tetramers are shown as green. Matrix protein depicted is from Newcastle disease virus (PDB 4g1g). Viral glycoproteins are based on PIV5 (PDB: 4gip for the F protein; PDB ID: 4jf7 and 3tsi for the PIV5 HN ectodomain and stalk, respectively. The vesicular stomatitis virus L protein structure (yellow) was used to represent the unknown paramyxovirus L conformation (PDB ID: 5a22). The phosphoprotein (brown) was modeled using the oligomerization domain of measles P (PDB ID: 3zdo). B) A model of an MeV virion in which a matrix protein array is located at the inner leaflet of the viral membrane. The matrix protein is shown in blue. The matrix-coated nucleocapsids were created using Chimera [92] using electron density maps EMD-1973 and EMD-1974 [43]. PDB structures were created in PyMOL [93].