Figure 4.
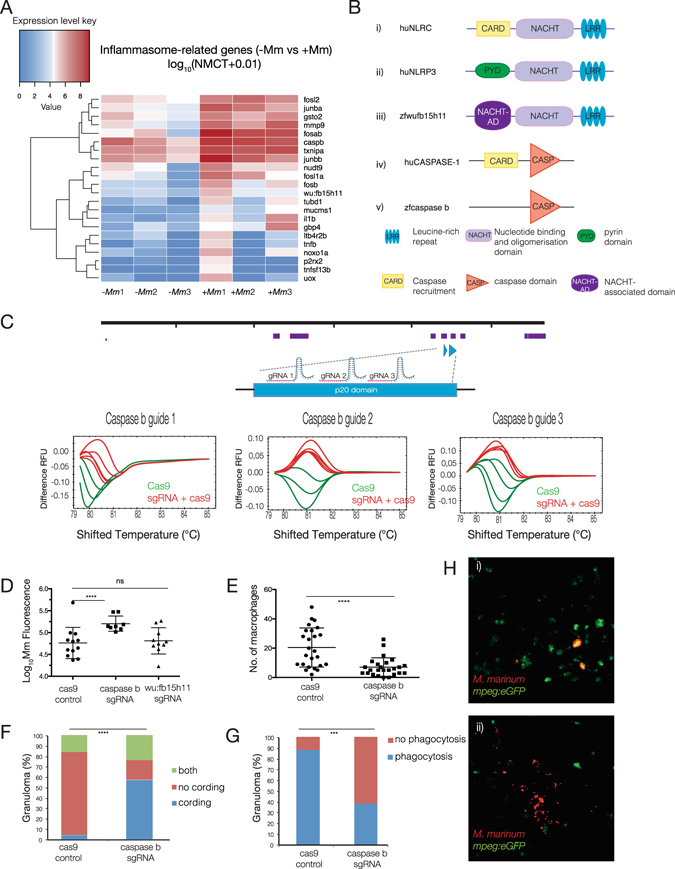
Neutrophils mediate M. marinum killing by inflammasome-dependent mechanism. (A) Heatmap shows the log10 (normalised counts (NMCT) +0.01) of selected differentially expressed transcripts of known inflammasome components and related genes (adjusted p-value < 0.05). Red - high expression. White - medium expression. Blue - low expression. (B) Schematic representation of the domain organisation of huNLRP3 (i), huNLRC (ii), zfwu:fb15h11 (iii), huCaspase 1 (iv) and zfcaspase b (v). (C) caspase b sgRNA tests for efficiency in inducing DNA double-strand breaks. sgRNAs were injected into the 1-cell stage and genomic DNA extracted at 24 hpf. High resolution melting curves from 3–4 individual embryos injected either with Cas9 mRNA only (green) or co-injected with Cas9 mRNA and caspase b sgRNA (red) are shown. (D–H) For quantitative analysis of inflammasome components, embryos were injected at the one-cell stage with Cas9 mRNA or sgRNAs to caspase b or wu:fb15h11 followed by injection of M. marinum at the 32–512 cell stage. (D) Bacterial burden of larvae infected at 3 dpi as measured by Mm fluorescence intensity (red). Statistical significance was determined by two-tailed unpaired students t-test with Welch’s correction. For caspase b sgRNA, ****p < 0.0001. (E) Enumeration of macrophage infiltrating M. marinum at 5 dpi. Statistical significance was determined by two-tailed unpaired students t-test with Welch’s correction (****p < 0.0001). (F) Percentage of larvae with cording phenotype at 5 dpi. Statistical significance was determined by chi-squared test (****p < 0.0001). (G) percentage of larvae where macrophages phagocytise M. marinum at 5 dpi. Statistical significance was determined by Fishers exact test (***p < 0.001). (H) Representative maximum intensity projections of larvae with cording bacteria with decreased phagocytosis in caspase b knockout embryos (i) vs non-cording controls (ii).
