Figure 6.
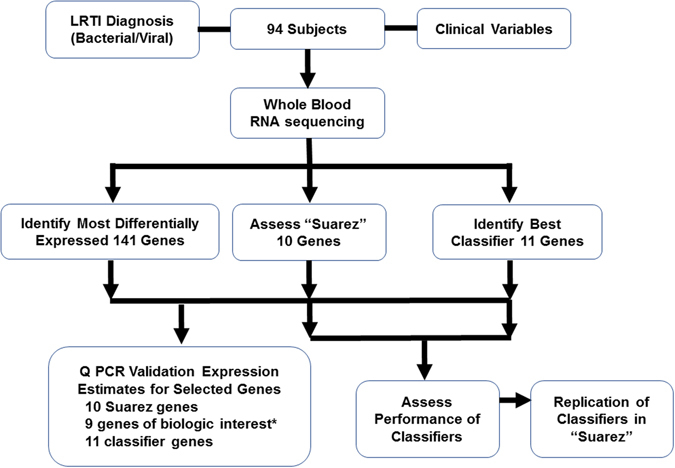
Flow chart of the analysis of different gene sets derived from RNA sequencing data from 94 subjects with confirmed bacterial-involved or non-bacterial LRTI selected for RNAseq interrogation. We first assessed expression of 10 genes identified by Suarez et al. to be differentially expressed comparing bacterial to viral infections. We next identified the 141 most differentially-expressed genes, as defined by statistical differences in bacterial vs. non-bacterial LRTI. Separately, we used a pathway-based approach to develop a novel gene expression classifier for discriminating bacterial vs. non-bacterial LRTI. *An additional 9 genes of biologic interest were selected from the 141gene set after considering the 10 Suarez genes. Validation of RNAseq-based expression estimates for selected genes, of particular biological interest, was attempted by qPCR. Finally, performance of the novel 3 pathway-based 11 gene classifier was assessed in our cohort.
