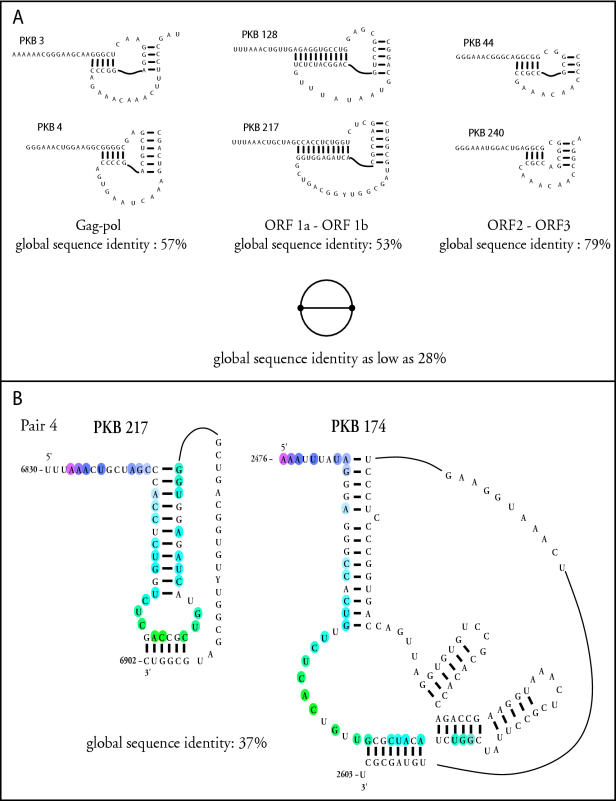
Figure 4.
Sequence and structural comparisons of pseudoknots in the frameshifting functional families. (A) Two secondary structures and corresponding topologies for each of the three viral frameshifting subfamilies Gag-pro, ORF1a/ORF1b and ORF2/ORF3. Sequence identity within the same subfamily ranges between 50 and 80%; it is as low as 28% when comparing pseudoknots from different subfamilies. All frameshifting pseudoknots are functionally related and have the same topology (dual graph). (B) Secondary structures and their aligned nucleotides (color coded) for frameshifting pseudoknots of LDV (PKB217) and Rous sarcoma virus (PKB174). Global sequence alignments were performed using the program ALIGN with default (−16/−4) gap opening/gap extension penalties.