Fig. 1. The splicing variants (SVs) and haplotypes of A3H.
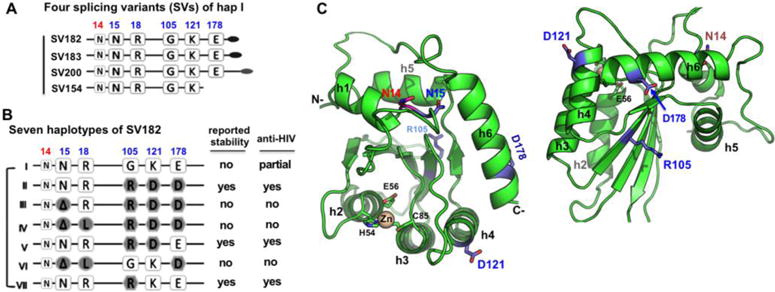
(A) Schematic overview of the four splicing variants based on haplotype I (hap I), with varying C-terminal extension. (B) The sequence variation of the seven haplotypes at the haplotype position 15, 18, 105, 121, and 178 based on SV182. The position 14 (colored in red), a conserved N in all A3H variants, is also shown. The reported stability inside cells and the anti-HIV activity for each haplotypes are listed on the right side based on literature. The weak anti-HIV activity of hap I is reported based on cell culture studies (10,11,17,18,27). (C) Two views of a modeled A3H hap II structure based on A3C (PDB ID 3VOW), which, like several previously published A3H models (13,18,57), shares the same core fold with all the known APOBEC structures. The locations of the haplotype residues (in sticks) are mapped to the modeled A3H structure. N15 at the end of helix 1 (h1) is buried, while its neighboring residue N14 is exposed. The R105 is on δ4, the opposite side of h2, h3 and h4. D121 is at the beginning of h4, and D178 is near the end of h6.
