Figure 3.
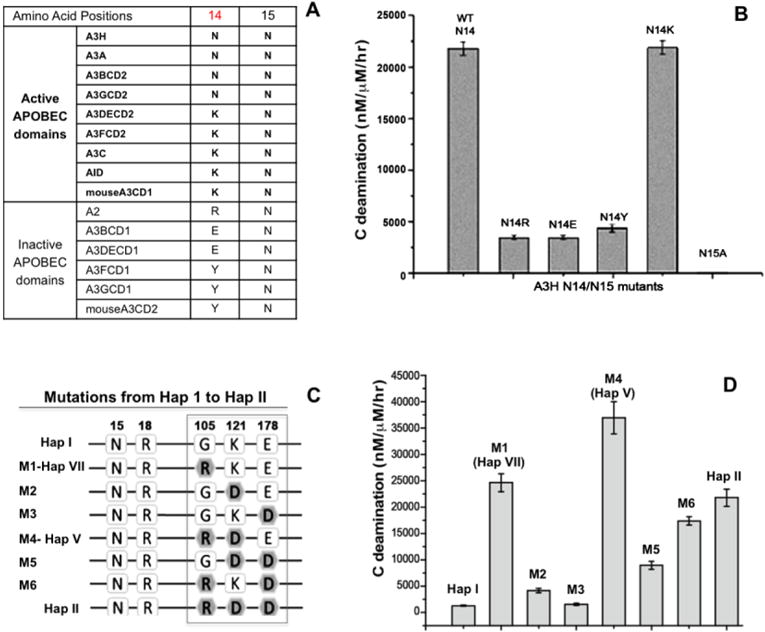
Mutational study to evaluate the effects of amino acids from the haplotypes on deaminase activity. (A) The sequence alignment at the residue number 14 and 15 of A3H with the active APOBEC domains and the inactive APOBEC domains. (B) The comparison of the deaminase activities of A3H mutants with substitutions at N14 and N15. The result indicates that position 14 with N14/K14 shows activity, but having the other three residues was about 10 times less active. For position 15, N15A mutation resulted in a complete loss of activity, similar to the ΔN15 mutant in hap III, IV, and VI. (C) Schematic overview showing the sequences at the haplotype residue position 105, 121 and 178 for hap I (first sequence) and hap II (last sequence), and the different combinations of mutations evolving from hap I to hap II. (D) The comparison of the deaminase activities of the different combination mutants are shown in panel-C. Error bars represent s.d. from the mean of three independent replicates.
