Figure 5.
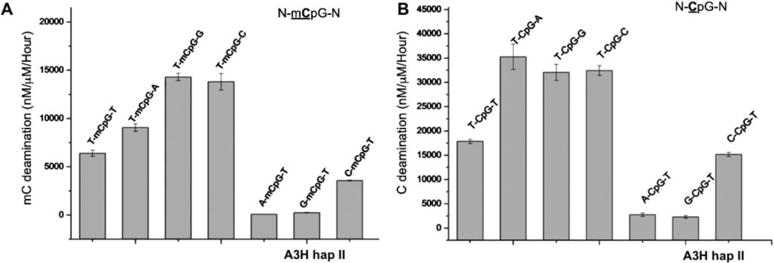
Examination of nucleotide sequence effect for deaminase using DNA substrate containing N-CpG-N or N-mCpG-N (the underlined C and mC are deamination target residue, and N being any nucleotide). (A) The deaminase activity of A3H hap II on the N-mC pG-N substrates, showing that T-mCpG-C/G are the best substrate. Error bars represent s.d. from the mean of three independent replicates. (B) The deaminase activity of A3H hap II on the N-CpG-N containing substrates, showing that T-CpG-C/G/A are the best substrates than others.
