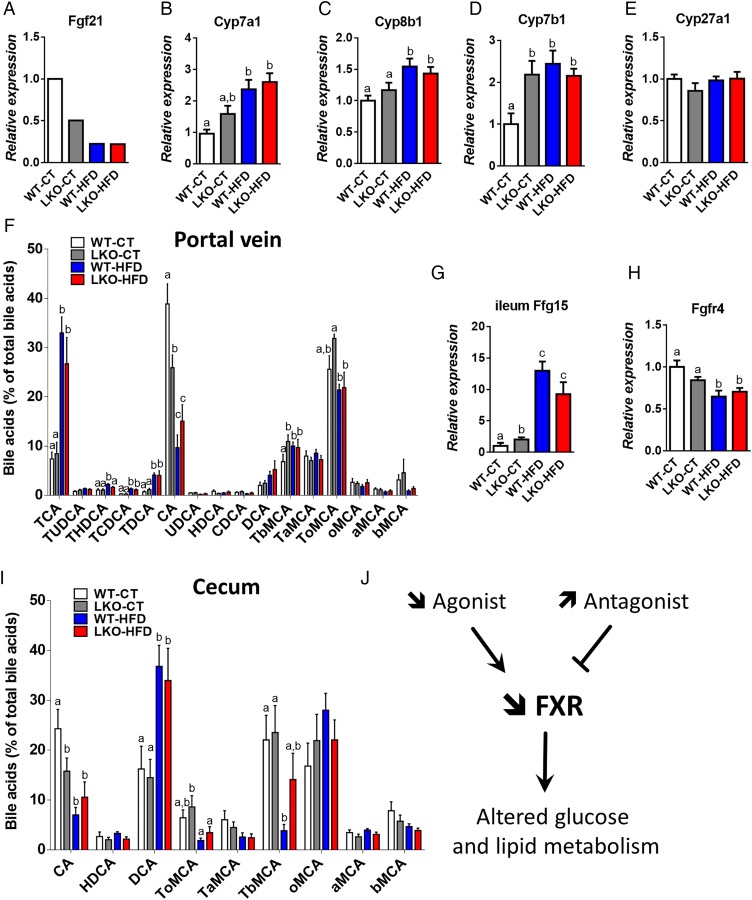
Figure 5.
Hepatocyte myeloid differentiation primary-response gene 88 (Myd88) deletion increases mRNA expression of genes involved in bile acid synthesis and signalling and affects plasma bile acid profile. (A) Fgf21 expression issued from the microarray analysis, (B) Cyp7a1 mRNA, (C) Cyp8b1 mRNA, (D) Cyp7b1 mRNA and (E) Cyp27a1 mRNA levels measured by qPCR in the liver of wild type (WT)-control diet (CT), liver-specific knockout (LKO)-CT, WT-high-fat diet (HFD) and LKO-HFD mice. These data correspond to the results of at least two independent experiments (n=12–20). Data are presented as mean±SEM. Data with different superscript letters are significantly different (p<0.05) according to post hoc one-way analysis of variance (ANOVA). (F) Plasma bile acids content (% of total bile acids), TCA, TUDCA, THDCA, TCDCA, TDCA, CA, UDCA, HDCA, CDCA, DCA, TbMCA, TaMCA, ToMCA, oMCA, aMCA, bMCA. Data are presented as mean±SEM (n=9–10). (G) Fgf15 mRNA in the ileum (n=12) and (H) Fgfr4 mRNA (n=10–20) in the liver of WT-CT, LKO-CT, WT-HFD and LKO-HFD. (I) Caecum bile acids content (% of total bile acids). Data are presented as mean±SEM (n=9–10). (J) Graphical summary: impact of a decreased abundance of FXR agonist and an increased abundance of FXR antagonist, leading to a decreased FXR activity and altered glucose, lipid metabolism. Data with different superscript letters are significantly different (p<0.05) according to post hoc one-way ANOVA. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; HDCA, hyodeoxycholic acid; LCA, lithocholic acid; MCA, muricholic acid; T, taurine; UDCA, ursodeoxycholic acid. a, alpha; b, beta; o, omega conjugated species.