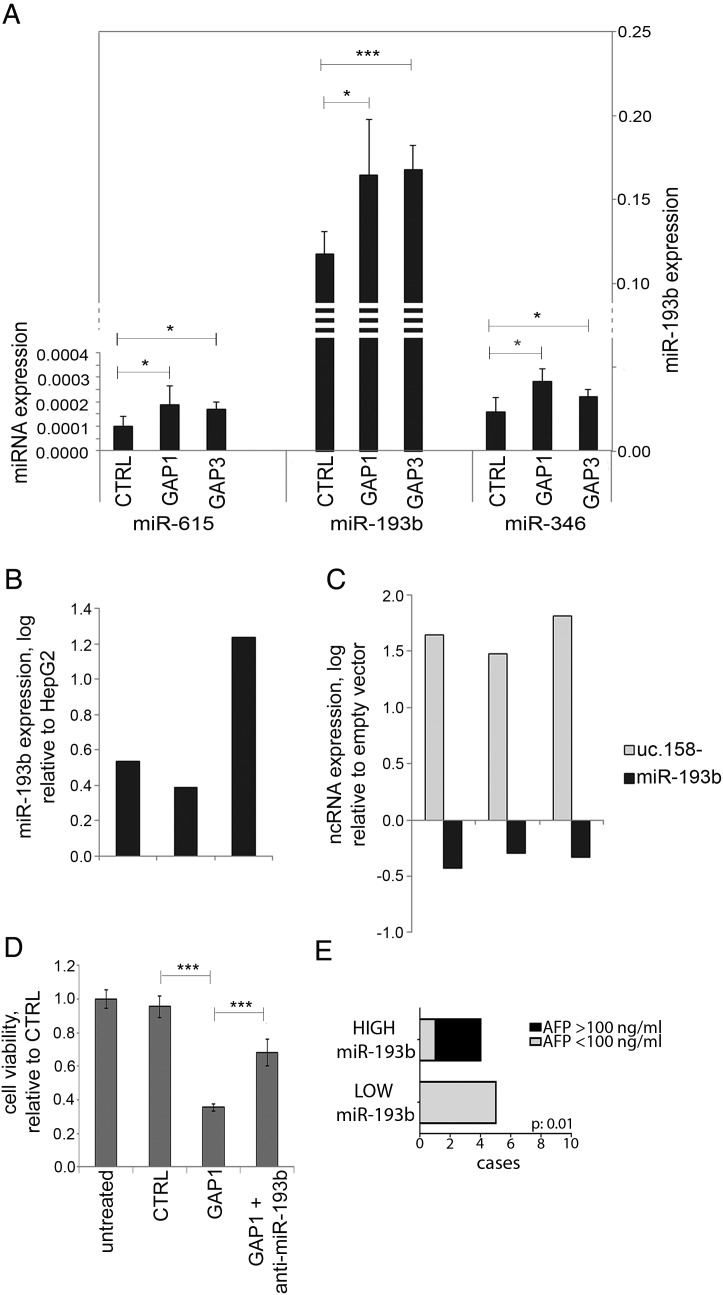
Figure 7.
miR-193b expression is dependent on uc.158− expression. HepG2 cells were transfected in 100 mm dishes for 48 hours, and RNA extracted to assess microRNA expression by TaqMan assays. Bars represent mean±SD of three experiments. (B) miR-193b expression was assessed in Huh-7 and HepG2 cells. Bars represent logarithmic expression of miR-193b in Huh-7 versus HepG2. In three independent experiments, miR-193b expression was higher in Huh-7 in comparison with HepG2. (C) Huh-7 cells were transfected with a vector overexpressing uc.158− or an empty vector for 48 hours. Expression of uc.158− and miR-193b was assessed by real-time PCR. Bars represent the logarithmic expression of ncRNAs in cells transfected with uc-158− vector compared with empty vector. The downregulation of mR-193 is quantitatively comparable with the upregulation achieved in panel A. (D) HepG2 cells were transfected with GAPMER control or GAP1 alone or in association with anti-miR-193b, and cell viability assessed by CellTiter-Blue at 48 hours. Bars represent mean±SD of six replicates. (E) Sera were collected from patients with hepatocarcinoma (HCC) naïve for any treatment (n:10), and microRNA extracted using fixed starting volume. miR-193b was assessed by TaqMan assay. χ2 test was used to assess the correlation between miR-193b and alpha-fetoprotein (AFP) groups.