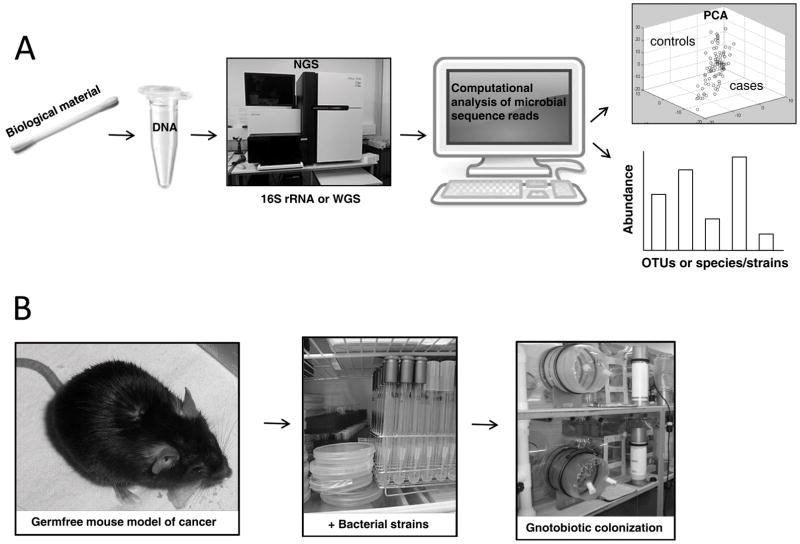
Figure 1. Microbiome research strategy.
(A) Flow chart of metagenomic sequence analysis. Biological material (buccal swabs, fecal samples, tissue biopsies, saliva) are procured from disease cases and healthy controls (panel 1); DNA is prepared from each sample (panel 2); Next-generation DNA sequencing (NGS) is performed to obtain targeted (16S rRNA hypervariable regions) or whole-genome shotgun (WGS) sequence reads (panel 3); Computational assembly and analysis of microbial sequence reads allows the microbial community structure to be assessed for each sample (panel 4); Principal Component Analysis (PCA) is a statistical procedure that compares the degree of relatedness of sequence reads between samples and illustrates the relationship between cases (red circles) and controls (blue circles), which often form distinct clusters with minimal overlap (panel 5 upper). Other computational methods allow the abundance of different microbial taxa to be quantified when compared to databases (panel 5 lower). Analysis of 16S data yields the relative abundance of Operational Taxanomic Units (OTUs) and their phylogenetic relationships. Analysis of WGS data provides greater taxonomic resolution, down to the abundance of specific strains within a single species that vary with respect to gene content including virulence factors and single nucleotide polymorphisms (SNPs), and provides more insight into pathways. WGS provides much more information but is more expensive and computationally intensive with less complete database resources, in part, due to a limited number of reference genomes. Further details can be found in other reviews (e.g., 9, 11). (B) Because a microbiome change between cases and controls can be either a cause or consequence of disease, gnotobiotic mouse models are utilized to evaluate the function of specific microbiota in the host. Germfree mouse models, which were originally obtained via C-section delivery but are now obtained by embryo transfer into germfree surrogate females (panel 1), are colonized by oral gavage (panel 2) with one bacterial strain (monoassociated), a consortium of specific bacteria (polyassociated), or complex microbial communities (e.g., fecal microbiota transplants) while maintained in gnotobiotic isolators (panel 3).