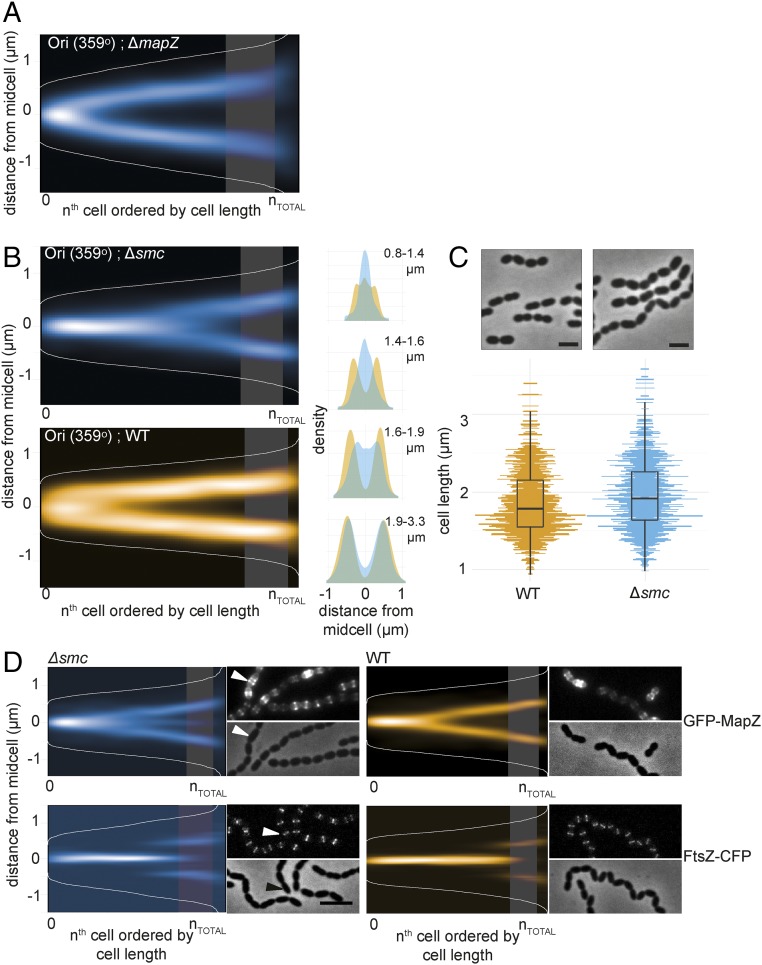
Fig. 4.
SMC is required for origin segregation and accurate division site selection. (A) Localization of the chromosomal origin (ParBp-GFP/parSp at 359°) in a ΔmapZ background shows that MapZ does not affect origin segregation (RR99). The data represent 2,976 cells/7,020 localizations. The shaded areas in all density histograms in this figure represent the point in wild-type cells where 50% of FtsZ localizes to the 1/4 positions of the cell. (B) The origins (ParBp-GFP/parSp at 359°) are segregated at a later stage in the cell cycle in Δsmc compared with wild type. The localizations are shown as heatmaps when cells are sorted according to length (Left) and as overlay of both localization density plots when the cells are grouped in four quartiles by cell length (Right). The data represent 2,012 cells/3,815 localizations for wild-type (MK359) and 3,908 cells/5,192 localizations for the Δsmc mutant (MK368). (C) Phase contrast images of wild-type D39 and Δsmc cells (AM39). (Scale bar, 2 µm.) Comparison of cell lengths between the wild type (1,407 cell analyzed; Top) and Δsmc (1,035 cells analyzed; Bottom). (D) Localization of GFP-MapZ and FtsZ-CFP in wild type versus Δsmc. Fluorescence and phase contrast micrographs are shown along with heatmaps. (Scale bar, 3 μm.) The arrowhead in the micrograph points to a cell with clearly mislocalized MapZ. Data represent 2,560 cells/5,314 localizations (∆smc, GFP-MapZ, RR110), 1,300 cells/2,257 localizations (∆smc, FtsZ-CFP, RR84), 3,908 cells/3,128 localizations (wild type, GFP-MapZ, RR101), and 3,422 cells/29,464 localizations (wild type, FtsZ-CFP, RR70).