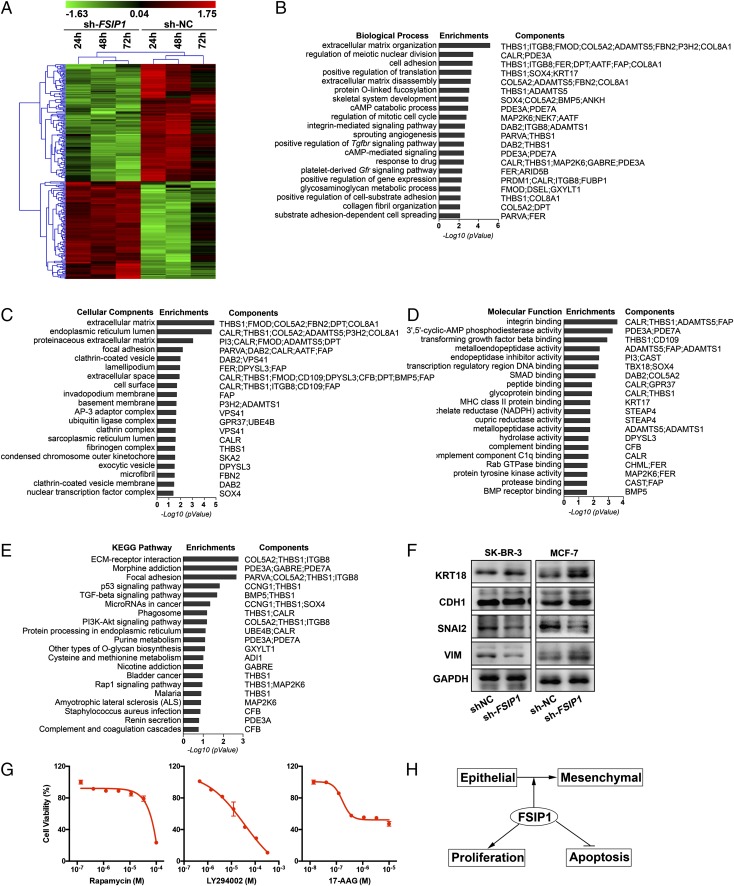
Fig. 5.
FSIP1 inhibition tracks with extracellular matrix components. (A) Microarray analysis (Affymetrix) of mRNA extracted from MCF-7 cells transfected with sh-NC or sh-FSIP1 for 24, 48 and 72 h, shown as a heat map of regulated genes (gene clusters noted; Materials and Methods). (B–E) A gene signature was derived from this dataset (Dataset S1), which was used to interrogate the GO (B–D) or KEGG (E) databases. The GO terms included biological processes, cellular components, and molecular functions. Note that the top hit in both GO and KEGG related to extracellular matrix proteins. (F) Western immunoblot confirmation of the expression of extracellular matrix interacting gene products involved in epithelial–mesenchymal transition, namely KRT18, CDH1, SNAI2, and vimentin (VIM) (GAPDH as loading control), in SKBR3 and MCF-7 cells transfected with sh-NC or sh-FSIP1 . (G) Effect of the top three hits obtained on CMAP interrogation (Table S1)—rapamycin, 17-AAG, and LY294002—on cell viability after treatment for 3 d (MTT assay). (H) Schematic showing that FSIP1 induces epithelial–mesenchymal transition, accelerates proliferation, and inhibits apoptosis, accounting for the increased aggressiveness of FSIP1-positive breast cancers noted in humans (5).